6F1Q
 
 | |
6EGZ
 
 | | Crystal structure of cytochrome c in complex with di-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SODIUM ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
6EGY
 
 | | Crystal structure of cytochrome c in complex with mono-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
6ECJ
 
 | |
6DUJ
 
 | | Crystal structure of A51V variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Lei, H, Bowler, B.E. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82202721 Å) | | Cite: | Naturally Occurring A51V Variant of Human CytochromecDestabilizes the Native State and Enhances Peroxidase Activity.
J.Phys.Chem.B, 123, 2019
|
|
6CUN
 
 | |
6CUK
 
 | |
6C4W
 
 | |
6BTM
 
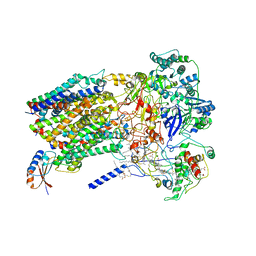 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
5ZKV
 
 | | Solution structure of molten globule state of L94G mutant of horse cytochrome-c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Naiyer, A, Islam, A, Hassan, M.I, Sundd, M, Ahmad, F. | | Deposit date: | 2018-03-26 | | Release date: | 2019-05-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of molten globule state of L94G mutant of horse cytochrome-c
To Be Published
|
|
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5XEC
 
 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5WVE
 
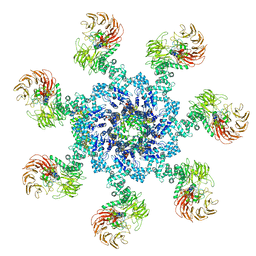 | | Apaf-1-Caspase-9 holoenzyme | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase, ... | | Authors: | Li, Y, Zhou, M, Hu, Q, Shi, Y. | | Deposit date: | 2016-12-24 | | Release date: | 2017-02-08 | | Last modified: | 2017-03-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TY3
 
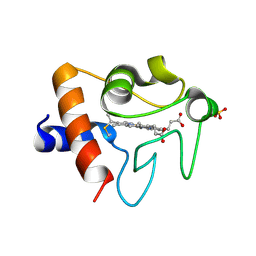 | | Crystal structure of K72A variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Mou, T.C, Nold, S.M, Lei, H, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effect of a K72A Mutation on the Structure, Stability, Dynamics, and Peroxidase Activity of Human Cytochrome c.
Biochemistry, 56, 2017
|
|
5T8W
 
 | |
5T7H
 
 | | Crystal structure of dimeric yeast iso-1-cytochrome C with CYMAL6 | | Descriptor: | 6-cyclohexylhexan-1-ol, Cytochrome c iso-1, HEME C, ... | | Authors: | Mcclelland, L, Mou, T.C, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-09-05 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Cytochrome c Can Form a Well-Defined Binding Pocket for Hydrocarbons.
J. Am. Chem. Soc., 138, 2016
|
|
5OCF
 
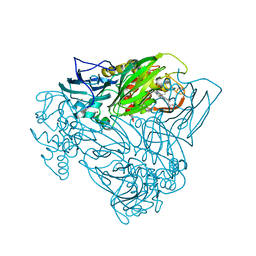 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OBO
 
 | | Crystal structure of nitrite bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, GLYCEROL, HEME C, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-28 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5O10
 
 | | Y48H mutant of human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Moreno-Chicano, T, Deacon, O.M, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Heightened Dynamics of the Oxidized Y48H Variant of Human Cytochrome c Increases Its Peroxidatic Activity.
Biochemistry, 56, 2017
|
|
5NCV
 
 | | Crystal Structure of Cytochrome c in complex with p-Methylphosphonatocalix[4]arene | | Descriptor: | CHLORIDE ION, Cytochrome c iso-1, HEME C, ... | | Authors: | Alex, J.M, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphonated Calixarene as a ""Molecular Glue"" for Protein Crystallization
Cryst.Growth Des., 18, 2018
|
|
5N1T
 
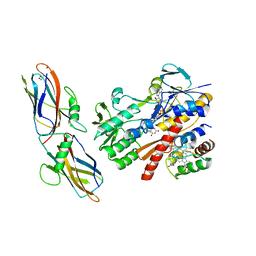 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | Descriptor: | COPPER (II) ION, CopC, Cytochrome C, ... | | Authors: | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LYC
 
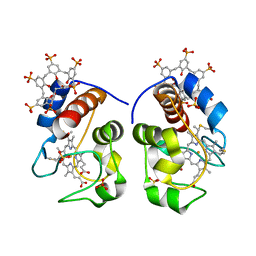 | |
