1W9B
 
 | | S. alba myrosinase in complex with carba-glucotropaeolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBA-GLUCOTROPAEOLIN, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The glucosinolate-myrosinase system. New insights into enzyme-substrate interactions by use of simplified inhibitors.
Org. Biomol. Chem., 3, 2005
|
|
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
1W9D
 
 | | S. alba myrosinase in complex with S-ethyl phenylacetothiohydroximate- O-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Glucosinolate-Myrosinase System. New Insights Into Enzyme-Substrate Interactions by Use of Simplified Inhibitors
Org.Biomol.Chem., 3, 2005
|
|
3CMJ
 
 | |
7F3A
 
 | | Arabidopsis thaliana GH1 beta-glucosidase AtBGlu42 | | Descriptor: | Beta-glucosidase 42, GLYCEROL | | Authors: | Horikoshi, S, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate specificity of glycoside hydrolase family 1 beta-glucosidase AtBGlu42 from Arabidopsis thaliana and its molecular mechanism.
Biosci.Biotechnol.Biochem., 86, 2022
|
|
5IDI
 
 | | Structure of beta glucosidase 1A from Thermotoga neapolitana, mutant E349A | | Descriptor: | 1,4-beta-D-glucan glucohydrolase, ACETATE ION | | Authors: | Kulkarni, T, Nordberg Karlsson, E, Logan, D.T. | | Deposit date: | 2016-02-24 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-glucosidase 1A from Thermotoga neapolitana and comparison of active site mutants for hydrolysis of flavonoid glucosides.
Proteins, 85, 2017
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4V
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
6RJK
 
 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | Descriptor: | Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RK2
 
 | | Complex structure of virulence factor SghA mutant with its substrate SAG | | Descriptor: | 2-(alpha-L-altropyranosyloxy)benzoic acid, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJM
 
 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | Descriptor: | Beta-glucosidase, alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJO
 
 | | Complex structure of virulence factor SghA with its substrate analog salicin | | Descriptor: | 2-(hydroxymethyl)phenyl beta-D-glucopyranoside, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-28 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7Z1I
 
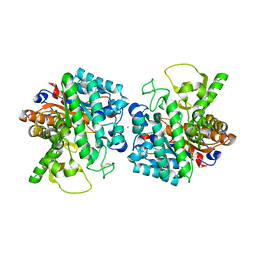 | | Plant myrosinase TGG1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Farmer, E, Jimenez-Sandoval, P, Santiago, J. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Plant myrosinase TGG1 from Arabidopsis thaliana
To Be Published
|
|
8HT0
 
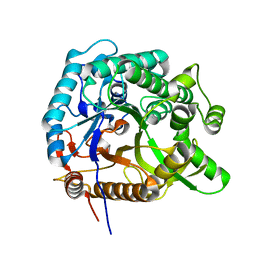 | |
6R4K
 
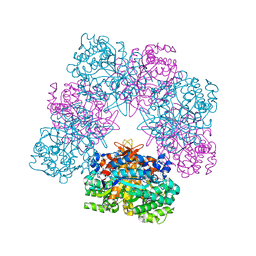 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with a monovalent inhibitor | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-[2-[2-(2-azanylidenehydrazinyl)ethoxy]ethoxy]phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
8IVY
 
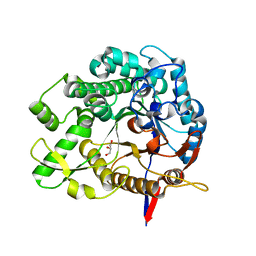 | | Beta-Glucosidase BglA mutant E166Q in complex with glucose | | Descriptor: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | Authors: | Dong, S, Xiao, Y, Feng, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Key roles of beta-glucosidase BglA for the catabolism of both laminaribiose and cellobiose in the lignocellulolytic bacterium Clostridium thermocellum.
Int.J.Biol.Macromol., 250, 2023
|
|
6QWI
 
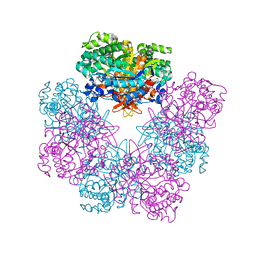 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with multivalent inhibitors. | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-(2-ethoxyethoxy)phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
8IN1
 
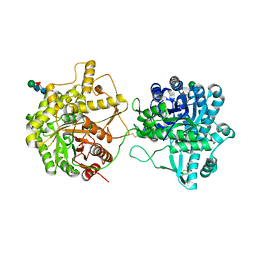 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8J3M
 
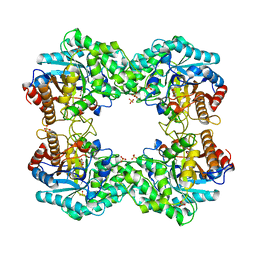 | |
8J5M
 
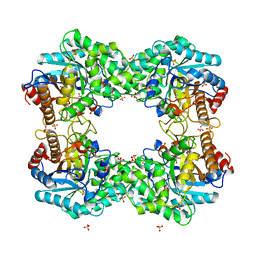 | |
8J5L
 
 | |
8B80
 
 | | The structure of Gan1D W433A in complex with galactose-6P | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of Gan1D W433A in complex with galactose-6P
To Be Published
|
|
8B81
 
 | | The structure of Gan1D W433A in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | The structure of Gan1D W433A in complex with cellobiose-6-phosphate
To Be Published
|
|
8XPC
 
 | | Crystal structure of Tris-bound TsaBgl (DATA I) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPD
 
 | | Crystal structure of Tris-bound TsaBgl (DATA II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
