2Y4U
 
 | |
2L6L
 
 | | Solution structure of human J-protein co-chaperone, Dph4 | | Descriptor: | DnaJ homolog subfamily C member 24, ZINC ION | | Authors: | Thakur, A, Chitoor, B.S, Atreya, H.S, Silva, P.D. | | Deposit date: | 2010-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanistic insights into novel iron-mediated moonlighting functions of human J-protein cochaperone, Dph4.
J.Biol.Chem., 287, 2012
|
|
7KAO
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAS
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAH
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAT
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAQ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAJ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAI
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAR
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAP
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KB5
 
 | |
7KAU
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
8J07
 
 | |
5NRO
 
 | | Structure of full-length DnaK with bound J-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaJ, Chaperone protein DnaK, ... | | Authors: | Kopp, J, Kityk, R, Mayer, M.P. | | Deposit date: | 2017-04-25 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones.
Mol. Cell, 69, 2018
|
|
5Y88
 
 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
6BYR
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
4WB7
 
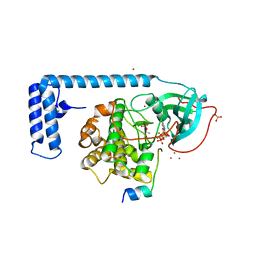 | | Crystal structure of a chimeric fusion of human DnaJ (Hsp40) and cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7Z3O
 
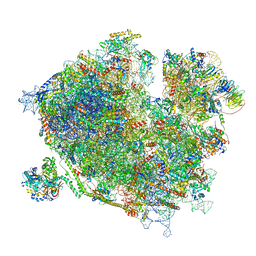 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-2 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3N
 
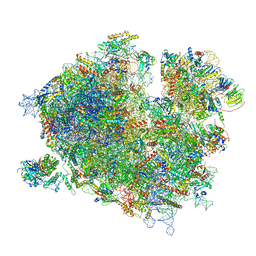 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6YXX
 
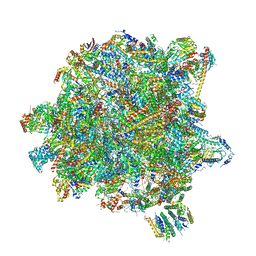 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
5AYK
 
 | | Crystal structure of ERdj5 form I | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
5AYL
 
 | | Crystal structure of ERdj5 form II | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
