5Y4D
 
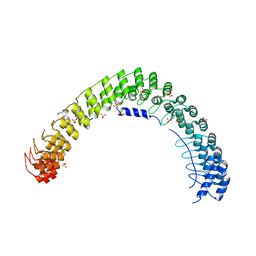 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | Descriptor: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4F
 
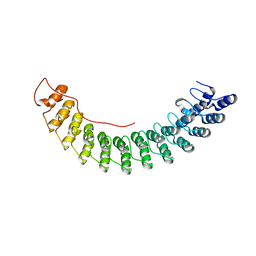 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5YBJ
 
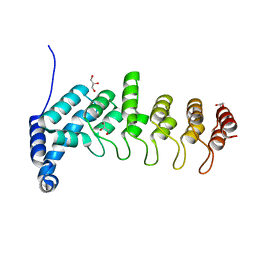 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
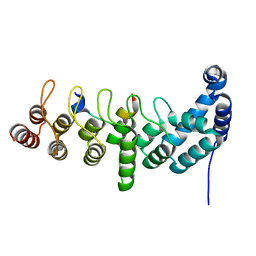 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5H2C
 
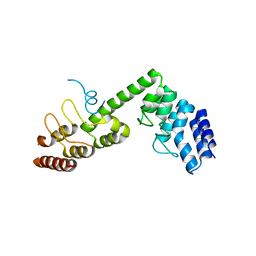 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | Descriptor: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H28
 
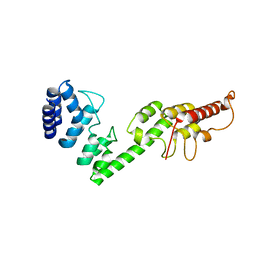 | |
5H2A
 
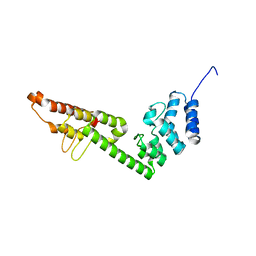 | |
3JXJ
 
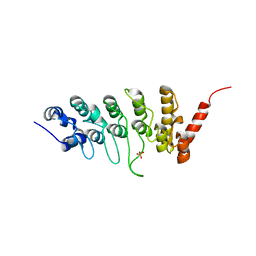 | |
5JHQ
 
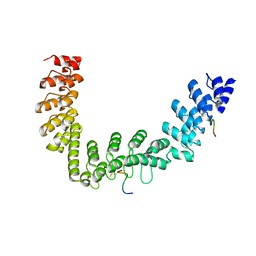 | |
3JXI
 
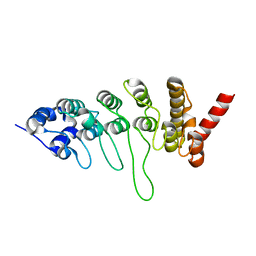 | |
3UXG
 
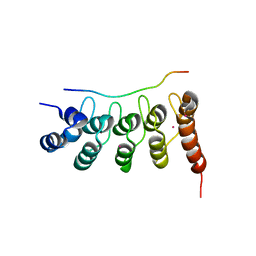 | | Crystal structure of RFXANK | | Descriptor: | DNA-binding protein RFXANK, Histone deacetylase 4, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chao, X, Bian, C, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3W9F
 
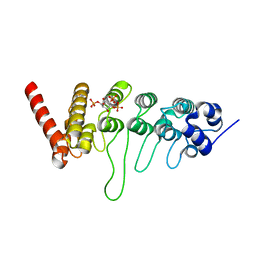 | | Crystal structure of the ankyrin repeat domain of chicken TRPV4 in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Vanilloid receptor-related osmotically activated channel protein | | Authors: | Itoh, Y, Hamada-nakahara, S, Suetsugu, S. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TRPV4 channel activity is modulated by direct interaction of the ankyrin domain to PI(4,5)P2
Nat Commun, 5, 2014
|
|
3W9G
 
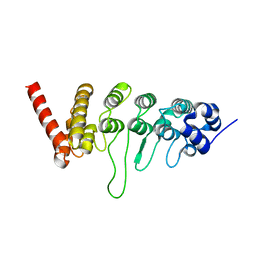 | |
3UI2
 
 | |
3V30
 
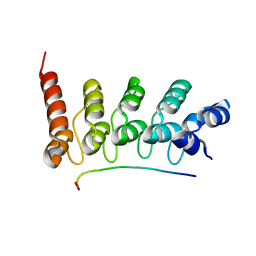 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
6MEW
 
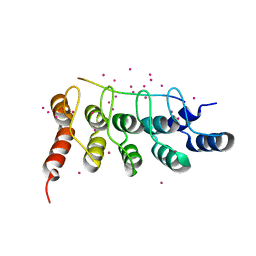 | | RFXANK ankyrin repeats in complex with a RFX7 peptide | | Descriptor: | DNA-binding protein RFXANK, RFX7 peptide, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | RFXANK ankyrin repeats in complex with a RFX7 peptide
to be published
|
|
5JA4
 
 | |
4N5Q
 
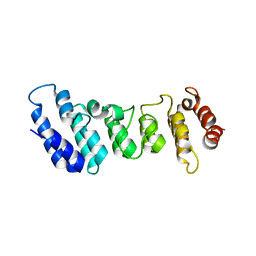 | | Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Shi, D.J, Ye, S, Cao, X, Wang, K.W, Zhang, R. | | Deposit date: | 2013-10-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function
Protein Cell, 4, 2013
|
|
2QC9
 
 | |
2FO1
 
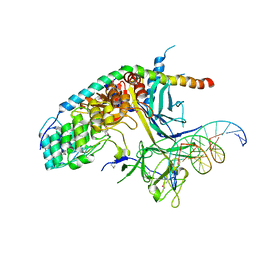 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
4F1P
 
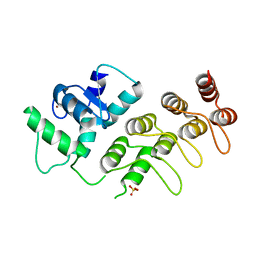 | | Crystal Structure of mutant S554D for ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2012-05-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
7T4X
 
 | |
4LG6
 
 | | Crystal structure of ANKRA2-CCDC8 complex | | Descriptor: | Ankyrin repeat family A protein 2, Coiled-coil domain-containing protein 8, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Bian, C, Tempel, W, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ankyrin Repeats of ANKRA2 Recognize a PxLPxL Motif on the 3M Syndrome Protein CCDC8.
Structure, 23, 2015
|
|
5VKQ
 
 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
6YSN
 
 | | Human TRPC5 in complex with Pico145 (HC-608) | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 5 | | Authors: | Wright, D.J, Johnson, R.M, Muench, S.P, Bon, R.S. | | Deposit date: | 2020-04-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site.
Commun Biol, 3, 2020
|
|
