6SQQ
 
 | |
6SQV
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/R70W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
3IRW
 
 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6SNJ
 
 | |
6SQT
 
 | |
4CH0
 
 | | RRM domain from C. elegans SUP-12 | | Descriptor: | PROTEIN SUP-12, ISOFORM B | | Authors: | Amrane, S, Mackereth, C.D. | | Deposit date: | 2013-11-27 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone-Independent Nucleic Acid Binding by Splicing Factor Sup-12 Reveals Key Aspects of Molecular Recognition
Nat.Commun., 5, 2014
|
|
5IM0
 
 | |
3IIN
 
 | | Plasticity of the kink turn structural motif | | Descriptor: | DNA/RNA (5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*GP*AP*CP*C)-D(P*AP*GP*A)-R(P*CP*GP*GP*CP*C)-3'), DNA/RNA (5'-R(*CP*A)-D(P*T)-3'), Group I intron, ... | | Authors: | Lipchock, S.V, Strobel, S.A, Antonioli, A.H, Cochrane, J.C. | | Deposit date: | 2009-08-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Plasticity of the RNA kink turn structural motif.
Rna, 16, 2010
|
|
6SQN
 
 | |
6SXW
 
 | | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638) | | Descriptor: | SULFATE ION, Zinc finger protein 638 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-09-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638)
To Be Published
|
|
6TR0
 
 | | Solution structure of U2AF2 RRM1,2 | | Descriptor: | Splicing factor U2AF 65 kDa subunit | | Authors: | Kang, H.-S, Sattler, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CIO
 
 | |
4CH1
 
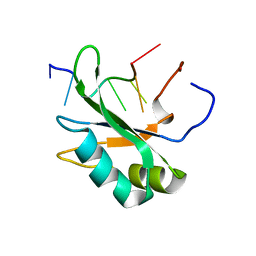 | |
5HO4
 
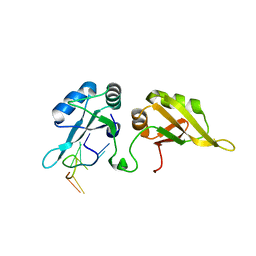 | | Crystal structure of hnRNPA2B1 in complex with 10-mer RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*AP*GP*C)-3') | | Authors: | Wu, B.X, Su, S.C, Gan, J.H, Ma, J.B. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5IQQ
 
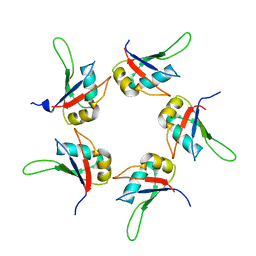 | |
3K0J
 
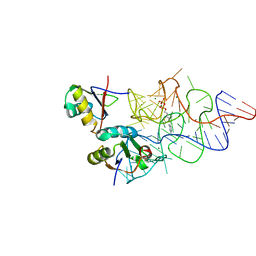 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
5ITH
 
 | |
3IWN
 
 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4ED5
 
 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4EGL
 
 | | Crystal structure of two tandem RNA recognition motifs of Human antigen R | | Descriptor: | ELAV-like protein 1, GLYCEROL, SULFATE ION | | Authors: | Wang, H, Zeng, F, Liu, H, Teng, M, Li, X. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of two tandem RNA recognition motifs of Human antigen R
To be Published
|
|
7ZIR
 
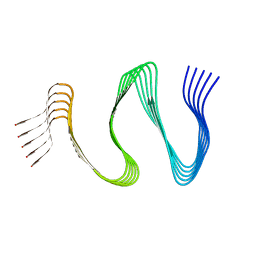 | | Cryo-EM structure of hnRNPDL amyloid fibrils | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like | | Authors: | Garcia-Pardo, J, Chaves-Sanjuan, A, Bartolome-Nafria, A, Gil-Garcia, M, Visentin, C, Bolognesi, M, Ricagno, S, Ventura, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3.
Nat Commun, 14, 2023
|
|
7ZUG
 
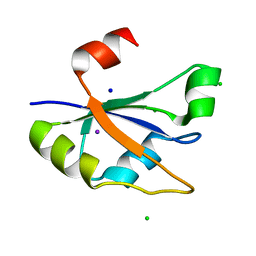 | | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain | | Descriptor: | CHLORIDE ION, Heterogeneous nuclear ribonucleoprotein H, N-terminally processed, ... | | Authors: | Winter, N, Kumar, M, Isupov, M.N, Wiener, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.075 Å) | | Cite: | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain
To Be Published
|
|
6CMN
 
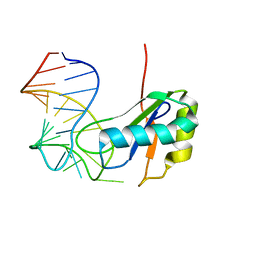 | | Co-Crystal Structure of HIV-1 TAR Bound to Lab-Evolved RRM TBP6.7 | | Descriptor: | TAR-Binding Protein 6.7, Trans-Activation Response RNA Element | | Authors: | Belashov, I.A, Wedekind, J.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structure of HIV TAR in complex with a Lab-Evolved RRM provides insight into duplex RNA recognition and synthesis of a constrained peptide that impairs transcription.
Nucleic Acids Res., 46, 2018
|
|
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
8B8S
 
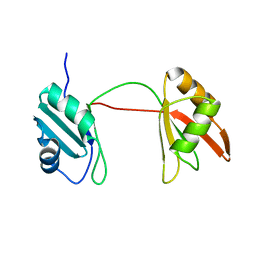 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-08 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
