1YE0
 
 | | T-To-T(High) quaternary transitions in human hemoglobin: betaV33A oxy (2MM IHP, 20% PEG) (1 test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, OXYGEN MOLECULE, ... | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-12-27 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
5YUI
 
 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y, McKenna, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5YUJ
 
 | |
2TMK
 
 | |
2XKG
 
 | | C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKI
 
 | | Aquo-met structure of C.lacteus mini-Hb | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKH
 
 | | Xe derivative of C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
3HUE
 
 | |
3HUF
 
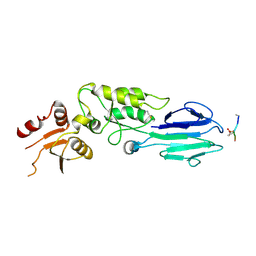 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
5WQD
 
 | |
6SQO
 
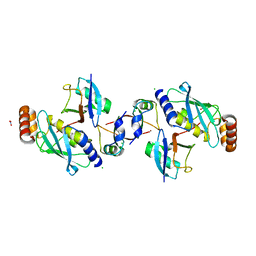 | | Crystal structure of human MDM2 RING domain homodimer bound to UbcH5B-Ub | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6SQS
 
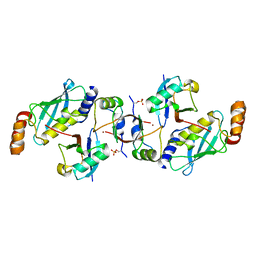 | | Crystal structure of cat phospho-Ser429 MDM2 RING domain bound to UbcH5B-Ub | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6SQR
 
 | | Crystal structure of Cat MDM2-S429E RING domain bound to UbcH5B-Ub | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
3MID
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (100mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MIH
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide, obtained in the presence of substrate | | Descriptor: | AZIDE ION, COPPER (II) ION, IODIDE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLK
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, NITRITE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIE
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (50mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLJ
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIG
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite, obtained in the presence of substrate | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLL
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide | | Descriptor: | AZIDE ION, COPPER (II) ION, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIB
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3O4Z
 
 | |
3MIC
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by co-crystallization | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
4A7N
 
 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4B1Y
 
 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
