6G1M
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
5EP9
 
 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EPA
 
 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIB
 
 | |
5GVJ
 
 | | Structure of FabK (M276A) mutant from Thermotoga maritima | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [FMN], SODIUM ION | | Authors: | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
4TMY
 
 | | CHEY FROM THERMOTOGA MARITIMA (MG-IV) | | Descriptor: | CHEY PROTEIN, MAGNESIUM ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
6TN6
 
 | | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | da Silva, V.M, Squina, F.M, Sperenca, M, Martin, L, Muniz, J.R.C, Garcia, W, Nicolet, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structure of a modular hyperthermostable endo-beta-1,4-mannanase from Thermotoga petrophila: The ancillary immunoglobulin-like module is a thermostabilizing domain.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
4C2L
 
 | | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-XYLOGALACTURONAN HYDROLASE A, ... | | Authors: | Rozeboom, H.J, Beldman, G, Schols, H.A, Dijkstra, B.W. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Endo-Xylogalacturonan Hydrolase from Aspergillus Tubingensis.
FEBS J., 280, 2013
|
|
2LQ8
 
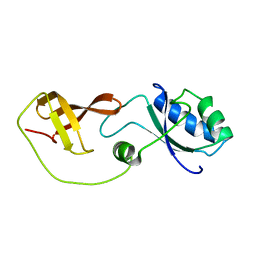 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
6W7S
 
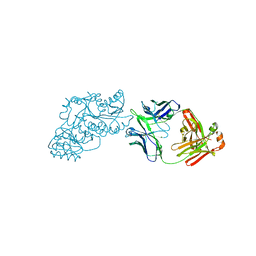 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 2G10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G10 (Fab heavy chain), 2G10 (Fab light chain), ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
4QDN
 
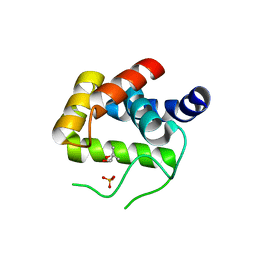 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
4ZRM
 
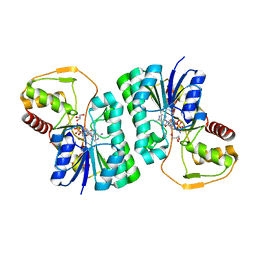 | |
4ZRN
 
 | |
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
2NSC
 
 | |
4GKL
 
 | |
4P6Y
 
 | |
5AN6
 
 | | Crystal structure of Thermotoga maritima Csm2 | | Descriptor: | CADMIUM ION, CRISPR-ASSOCIATED PROTEIN, CSM2 FAMILY | | Authors: | Gallo, G, Augusto, G, Rangel, G, Zelanis, A, Mori, M.A, Campos, C.B, Wurtele, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural Basis for Dimer Formation of the Crispr-Associated Protein Csm2 of Thermotoga Maritima.
FEBS J., 283, 2016
|
|
4I0U
 
 | | Improved structure of Thermotoga maritima CorA at 2.7 A resolution | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nordin, N, Guskov, A, Phua, T, Sahaf, N, Xia, Y, Lu, S.Y, Eshaghi, H, Eshaghi, S. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structure and function of Thermotoga maritima CorA reveals the mechanism of gating and ion selectivity in Co2+/Mg2+ transport.
Biochem.J., 451, 2013
|
|
4IGA
 
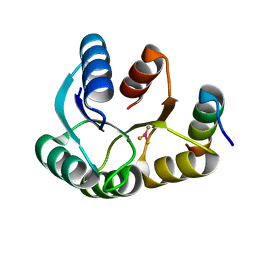 | |
2NSB
 
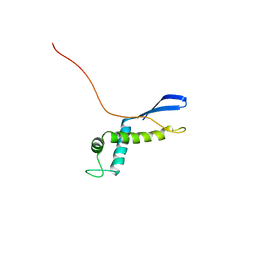 | |
2NSA
 
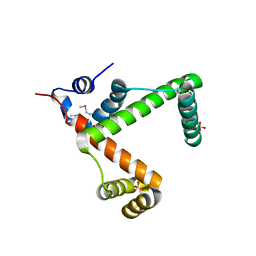 | |
3JUR
 
 | | The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima | | Descriptor: | Exo-poly-alpha-D-galacturonosidase | | Authors: | Pijning, T, van Pouderoyen, G, Kluskens, L.D, van der Oost, J, Dijkstra, B.W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of a hyperthermoactive exopolygalacturonase from Thermotoga maritima reveals a unique tetramer
Febs Lett., 2009
|
|
3TMY
 
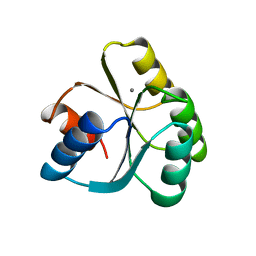 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
