5FRB
 
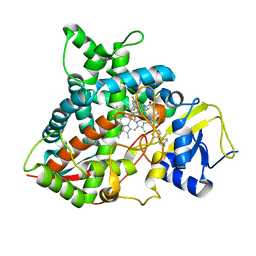 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with a tetrazole-based inhibitor VT-1598 | | Descriptor: | (R)-4-((4-((6-(2-(2,4-difluorophenyl)-1,1-difluoro-2-hydroxy-3-(1H-tetrazol-1-yl)propyl)pyridin-3-yl)ethynyl)phenoxy)methyl)benzonitrile, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2015-12-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of the New Investigational Drug Candidate VT-1598 in Complex with Aspergillus fumigatus Sterol 14 alpha-Demethylase Provides Insights into Its Broad-Spectrum Antifungal Activity.
Antimicrob. Agents Chemother., 61, 2017
|
|
4JC3
 
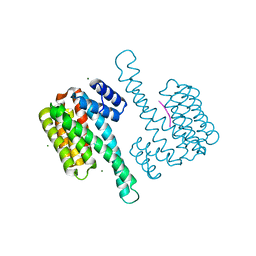 | |
6Y1J
 
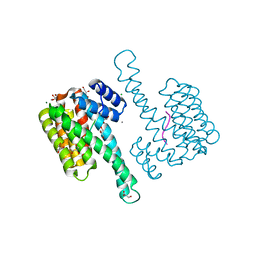 | |
6XWD
 
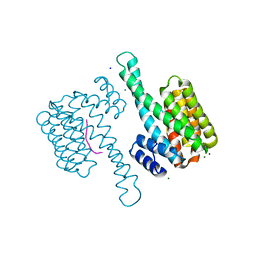 | |
7AEW
 
 | |
7AXN
 
 | | 14-3-3 sigma in complex with Pin1 binding site pS72 and covalently bound TCF521-026 | | Descriptor: | 14-3-3 protein sigma, 3-chloranyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AYF
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-110 | | Descriptor: | 14-3-3 protein sigma, 6-(2-bromanylimidazol-1-yl)pyridine-3-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AOG
 
 | | 14-3-3 sigma in complex with Pin1 binding site pS72 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6ZCJ
 
 | | 14-3-3sigma in complex with SLP76pS376 phosphopeptide crystal structure | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, SLP76pS376 | | Authors: | Soini, L, Leysen, S, Davis, J, Ottmann, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The 14-3-3/SLP76 protein-protein interaction in T-cell receptor signalling: a structural and biophysical characterization.
Febs Lett., 595, 2021
|
|
3L4D
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Leishmania infantum in complex with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-12-19 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate Preferences and Catalytic Parameters Determined by Structural Characteristics of Sterol 14{alpha}-Demethylase (CYP51) from Leishmania infantum.
J.Biol.Chem., 286, 2011
|
|
6YR6
 
 | | 14-3-3 sigma in complex with hDM2-186 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, ... | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR5
 
 | | 14-3-3 sigma in complex with hDMX-367 peptide | | Descriptor: | 14-3-3 protein sigma, Protein Mdm4, SULFATE ION | | Authors: | Wolter, M, Srdanovic, S, Ottman, C, Warriner, S, Wilson, A. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR7
 
 | | 14-3-3 sigma in complex with hDMX-342+367 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein Mdm4 | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
5OEG
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
5NWK
 
 | | 14-3-3c in complex with CPP and fusicoccin | | Descriptor: | 14-3-3 c-1 protein, FUSICOCCIN, PHOSPHOSERINE, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
5NWI
 
 | | 14-3-3c in complex with CPP | | Descriptor: | 14-3-3 c-1 protein, ACETATE ION, Potassium channel KAT1 | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
8EQ8
 
 | |
8EQH
 
 | |
8C3Z
 
 | |
8C40
 
 | | 14-3-3sigma bound to PKA-responsive ERa phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Estrogen Receptor alpha phosphopeptide, MAGNESIUM ION | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-12-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis and dual ligand regulation of tetrameric estrogen receptor alpha /14-3-3 zeta protein complex.
J.Biol.Chem., 299, 2023
|
|
6F08
 
 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
5OEH
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions. | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
5TZ1
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Candida albicans in complex with the tetrazole-based antifungal drug candidate VT1161 (VT1) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Hargrove, T, Wawrzak, Z, Lepesheva, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
3MHR
 
 | | 14-3-3 sigma in complex with YAP pS127-peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schumacher, B, Skwarczynska, M, Rose, R, Ottmann, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of a 14-3-3[sigma]-YAP phosphopeptide complex at 1.15 A resolution
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2O02
 
 | | Phosphorylation independent interactions between 14-3-3 and Exoenzyme S: from structure to pathogenesis | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, ExoS (416-430) peptide | | Authors: | Ottmann, C, Yasmin, L, Weyand, M, Hauser, A.R, Wittinghofer, A, Hallberg, B. | | Deposit date: | 2006-11-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis
Embo J., 26, 2007
|
|
