6R26
 
 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6UZK
 
 | |
2C83
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
5L93
 
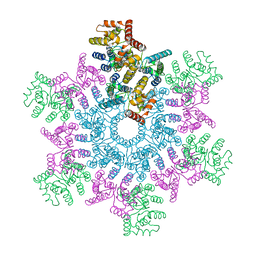 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
2C84
 
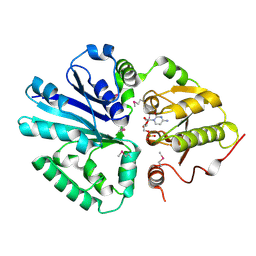 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
3COM
 
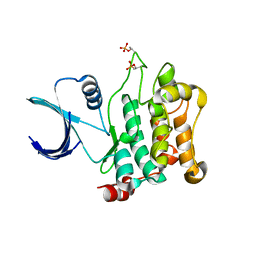 | | Crystal structure of Mst1 kinase | | Descriptor: | Serine/threonine-protein kinase 4 | | Authors: | Atwell, S, Burley, S.K, Dickey, M, Leon, B, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Mst1 kinase.
To be Published
|
|
2Z61
 
 | |
3EVK
 
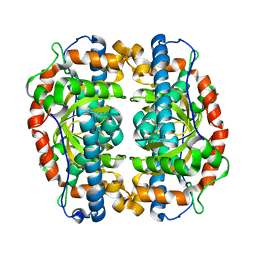 | |
4FZ5
 
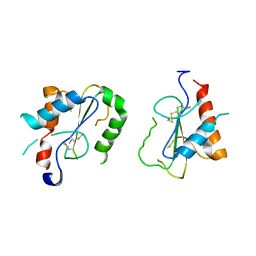 | | Crystal Structure of Human TIRAP TIR-domain | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Woo, J.R, Kim, S, Shoelson, S.E, Park, S. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray Crystallographic Structure of TIR-Domain from the Human TIR-Domain Containing Adaptor Protein/MyD88 Adaptor-Like Protein (TIRAP/MAL)
Bull.Korean Chem.Soc., 33, 2013
|
|
4HKD
 
 | |
4HQ6
 
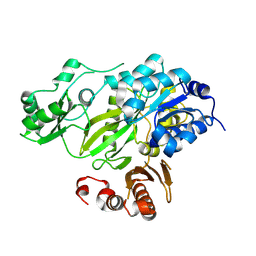 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
7W7Q
 
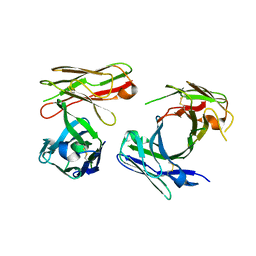 | |
1OAD
 
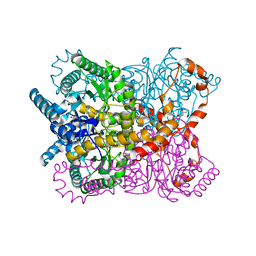 | | Glucose isomerase from Streptomyces rubiginosus in P21212 crystal form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramagopal, U.A, Dauter, M, Dauter, Z. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sad Manganese in Two Crystal Forms of Glucose Isomerase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2OKL
 
 | |
2WT7
 
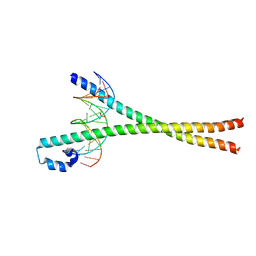 | | Crystal structure of the bZIP heterodimeric complex MafB:cFos bound to DNA | | Descriptor: | MODIFIED T-MARE MOTIF, PHOSPHATE ION, PROTO-ONCOGENE PROTEIN C-FOS, ... | | Authors: | Pogenberg, V, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-11 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
2NM0
 
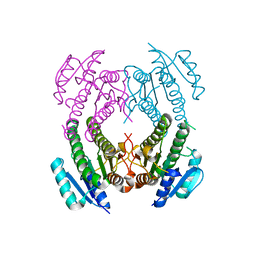 | | Crystal Structure of SCO1815: a Beta-Ketoacyl-Acyl Carrier Protein Reductase from Streptomyces coelicolor A3(2) | | Descriptor: | Probable 3-oxacyl-(Acyl-carrier-protein) reductase | | Authors: | Khosla, C, Tang, Y, Lee, H.Y, Tang, Y, Kim, C.Y, Mathews, I.I. | | Deposit date: | 2006-10-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2).
Biochemistry, 45, 2006
|
|
2WTY
 
 | | Crystal structure of the homodimeric MafB in complex with the T-MARE binding site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP *GP*CP*AP*AP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*TP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP *GP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Consani Textor, L, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
1R02
 
 | |
2N23
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the N-terminal activation domain of EKLF (TAD1) | | Descriptor: | Krueppel-like factor 1, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lecoq, L, Morse, T, Raiola, L, Arseneault, G, Omichinski, J. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Complex between the N-terminal Transactivation Domain of EKLF and the p62/Tfb1 subunit of TFIIH
To be Published
|
|
8FC4
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8G2C
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 2024
|
|
8G29
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 2024
|
|
8GN0
 
 | | Crystal structure of DCBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
