4FLQ
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLO
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLS
 
 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4AEF
 
 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | Descriptor: | NEOPULLULANASE (ALPHA-AMYLASE II) | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
4FLR
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290L from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4GI9
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with Trehalulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI8
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with the substrate sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GIN
 
 | | Crystal structure of the MUTB R284C mutant from crystals soaked with the inhibitor deoxynojirimycin | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI6
 
 | | Crystal structure of the MUTB F164L mutant in complex with glucose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2C
 
 | | Trehalulose synthase MutB R284C mutant | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GIA
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with isomaltulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GKL
 
 | |
4E2O
 
 | | Crystal structure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Mok, S.C, Teh, A.H, Saito, J.A, Najimudin, N, Alam, M. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans.
Enzyme.Microb.Technol., 53, 2013
|
|
3VU2
 
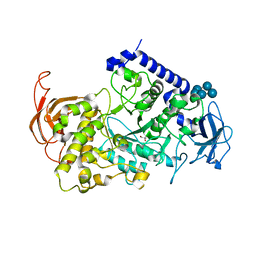 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltopentaose from Oryza sativa L | | Descriptor: | 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, GLYCEROL, ... | | Authors: | Chaen, K, Kakuta, Y, Kimura, M. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the rice branching enzyme I (BEI) in complex with maltopentaose.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
3VM7
 
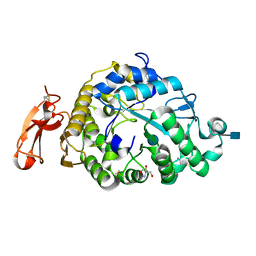 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
3ZOA
 
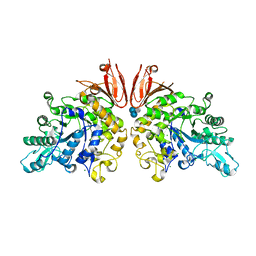 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
3ZO9
 
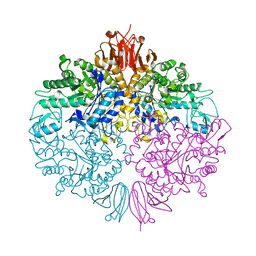 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
4GO9
 
 | | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTANT, MUTB D415N, in COMPLEX with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTB, MUTANT D415N, in COMPLEX with TRIS
To be Published, 2013
|
|
4GO8
 
 | | Crystal Structure of the TREHALULOSE SYNTHASE MUTB, MUTANT A258V, in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of MUTB A258V mutant in complex with TRIS
To be Published, 2013
|
|
4AYS
 
 | | The Structure of Amylosucrase from D. radiodurans | | Descriptor: | AMYLOSUCRASE | | Authors: | Skov, L.K, Pizzut, S, Remaud-Simeon, M, Gajhede, M, Mirza, O. | | Deposit date: | 2012-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Structure of Amylosucrase from Deinococcus Radiodurans Has an Unusual Open Active-Site Topology
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3VX1
 
 | |
3VX0
 
 | | Crystal Structure of alpha-amylase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase A type-1/2, CALCIUM ION, ... | | Authors: | Sugahara, M. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of alpha-amylase from Aspergillus oryzae
To be Published
|
|
4MB1
 
 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4MAZ
 
 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4M8U
 
 | | The Structure of MalL mutant enzyme V200A from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
