6XRM
 
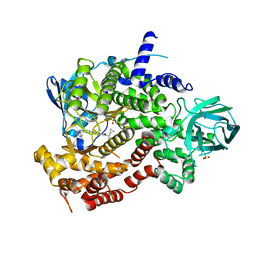 | | Crystal structure of human PI3K-gamma in complex with Compound 4 | | Descriptor: | 5-[2-amino-3-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]-2-[(1S)-1-cyclopropylethyl]-7-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XRN
 
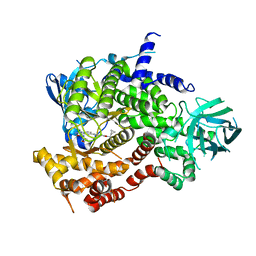 | | Crystal structure of human PI3K-gamma in complex with Compound 17 | | Descriptor: | 2-amino-5-{2-[(1S)-1-cyclopropylethyl]-7-methyl-1-oxo-2,3-dihydro-1H-isoindol-5-yl}-N-(trans-3-hydroxycyclobutyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Optimization of Pyrazolopyrimidine Amide Inhibitors of Phosphoinositide 3-Kinase gamma (PI3K gamma ).
J.Med.Chem., 65, 2022
|
|
6YKG
 
 | |
6Z2W
 
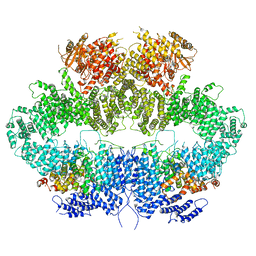 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z2X
 
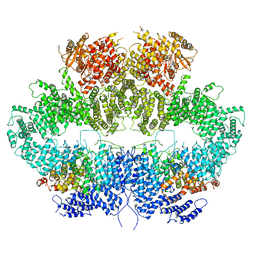 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP (State II) | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z3A
 
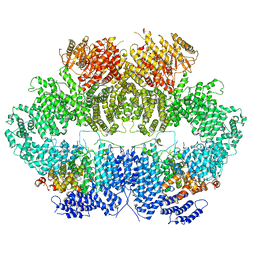 | | Mec1-Ddc2 (wild-type) in complex with AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase MEC1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z3R
 
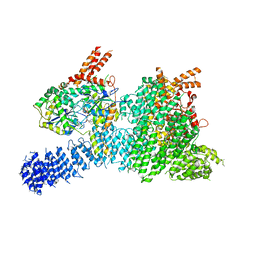 | | Structure of SMG1-8-9 kinase complex bound to UPF1-LSQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Gat, Y, Conti, E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity.
Elife, 9, 2020
|
|
6ZAA
 
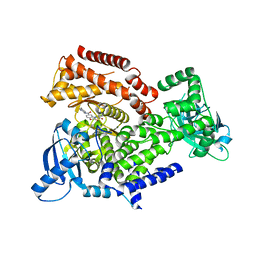 | | PI3K Delta in complex with methoxy(methylsulfamoyl)pyridinylN(methylpiperidinyl)dihydrobenzoxazinecarboxamide | | Descriptor: | 4-[6-methoxy-5-(methylsulfamoyl)pyridin-3-yl]-~{N}-(1-methylpiperidin-4-yl)-2,3-dihydro-1,4-benzoxazine-6-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-15 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZAC
 
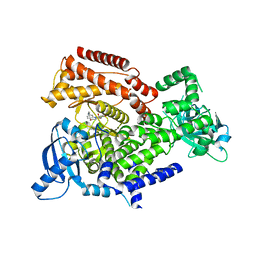 | | PI3K Delta in complex with [(dimethylamino)methyldihydrobenzoxazin2methoxypyridinyl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-[6-[(dimethylamino)methyl]-2,3-dihydro-1,4-benzoxazin-4-yl]-2-methoxy-pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZAD
 
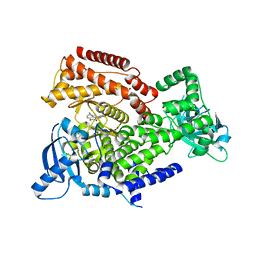 | | PI3K Delta in complex with methoxymethyloxathiatetraazatetracyclodocosahexaenedione | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methoxymethyloxathiatetraazatetracyclodocosahexaenedione | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-29 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZFP
 
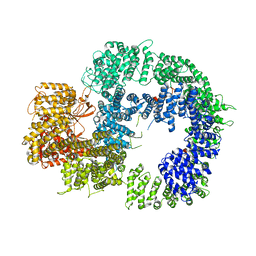 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
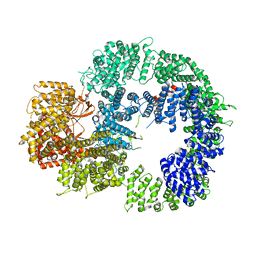 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH8
 
 | | Cryo-EM structure of DNA-PKcs:DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZWM
 
 | | cryo-EM structure of human mTOR complex 2, overall refinement | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6ZWO
 
 | | cryo-EM structure of human mTOR complex 2, focused on one half | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
7BI2
 
 | | PI3KC2aDeltaN and DeltaC-C2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, IODIDE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI4
 
 | | PI3KC2a core apo | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI6
 
 | | PI3KC2a core in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI9
 
 | | PI3KC2a core in complex with PIK90 | | Descriptor: | 1,2-ETHANEDIOL, N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BL1
 
 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
7JIS
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
