8JN8
 
 | |
8JN9
 
 | |
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQI
 
 | | GENE V PROTEIN MUTANT WITH ILE 47 REPLACED BY VAL 47 (I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
5J5K
 
 | | CRYSTAL STRUCTURE OF AFMP4P IN COMPLEX WITH PALMITIC ACID | | Descriptor: | PALMITIC ACID, Uncharacterized protein, alpha-D-mannopyranose | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2016-04-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel Class Of Virulence Factors In Penicillium Marneffei And Aspergillus Fumigatus Enhances Intracellular Survival In Monocytes By Arachidonic Acid Binding
To Be Published
|
|
3CHH
 
 | | Crystal Structure of Di-iron AurF | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8Y91
 
 | | Structure of NET-nomifensine in outward-open state | | Descriptor: | (4S)-2-methyl-4-phenyl-3,4-dihydro-1H-isoquinolin-8-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 2024
|
|
8Y8Z
 
 | |
8Y92
 
 | | structure of NET-Atomoxetine in outward-open state | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 2024
|
|
8YR2
 
 | |
4F1R
 
 | |
8Y95
 
 | | Structure of NET-NE in Occluded state | | Descriptor: | CHLORIDE ION, Noradrenaline, SODIUM ION, ... | | Authors: | Zhang, H, Xu, H.E, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 2024
|
|
5XAF
 
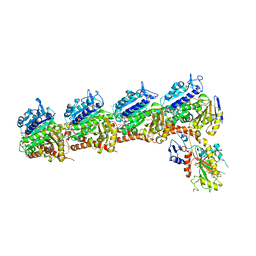 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
8Y93
 
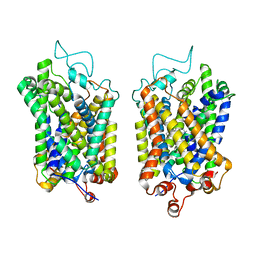 | |
8Y90
 
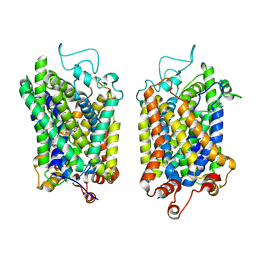 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 2024
|
|
2QZP
 
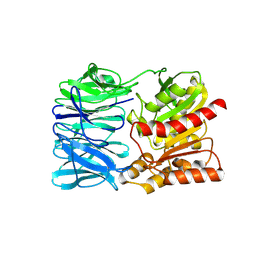 | |
8Y94
 
 | |
4F0V
 
 | |
4F0W
 
 | |
4F46
 
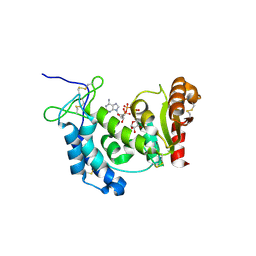 | | Crystal structure of wild type human CD38 in complex with NAADP and ADPRP | | Descriptor: | ADP-ribosyl cyclase 1, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5S)-3,4,5-tris(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2012-05-10 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structures of Human CD38 in Complex with NAADP and ADPRP
MESSENGER, 2, 2013
|
|
4F45
 
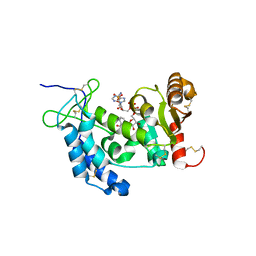 | | Crystal structure of human CD38 E226Q mutant in complex with NAADP | | Descriptor: | ADP-ribosyl cyclase 1, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2012-05-10 | | Release date: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human CD38 in Complex with NAADP and ADPRP
MESSENGER, 2, 2013
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
