9BE2
 
 | |
8CSH
 
 | |
3BTJ
 
 | |
3BTL
 
 | |
3KZ5
 
 | | Structure of cdomain | | Descriptor: | ACETATE ION, Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3TPT
 
 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPB
 
 | | Structure of HipA(S150A) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
2G66
 
 | |
6E4N
 
 | |
6E4O
 
 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
8DPK
 
 | |
4RS7
 
 | |
4S0R
 
 | | Structure of GS-TnrA complex | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N.G, Cuthbert, B, Tonthat, N.K. | | Deposit date: | 2015-01-04 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4RS8
 
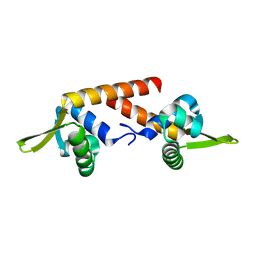 | |
1HAB
 
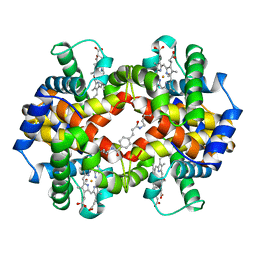 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1HAC
 
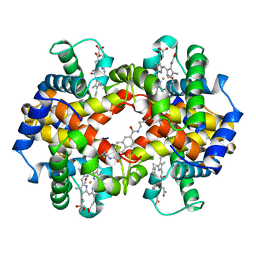 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6UEP
 
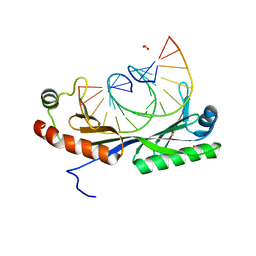 | |
6UEO
 
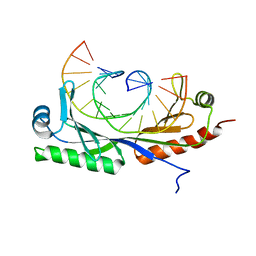 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
1WET
 
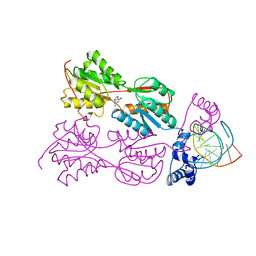 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
3VEA
 
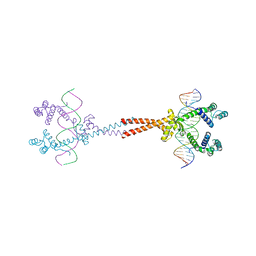 | | Crystal Structure of matP-matS23mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', Macrodomain Ter protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
3VEB
 
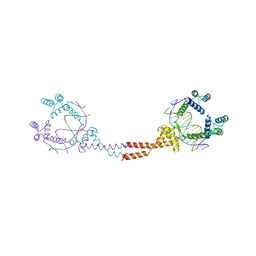 | | Crystal Structure of Matp-matS | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*G)-3', 5'-D(*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*G)-3', CALCIUM ION, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
3BTI
 
 | |
3BTC
 
 | |
