5DPA
 
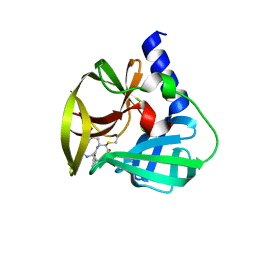 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5IDK
 
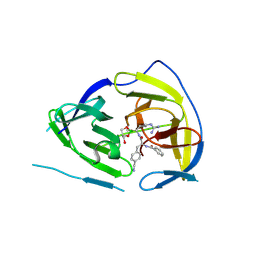 | |
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
4PV5
 
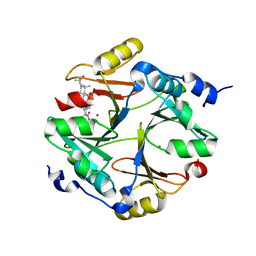 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHS
 
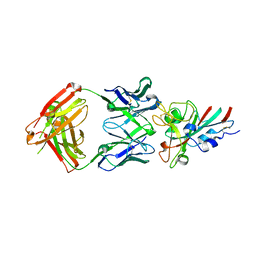 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
8J7B
 
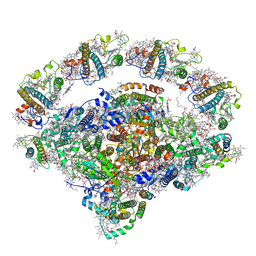 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 2 (PSI-ST2) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J6Z
 
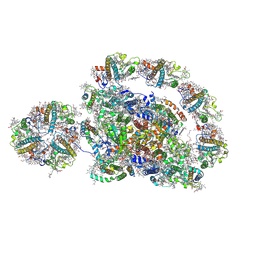 | | Cryo-EM structure of the Arabidopsis thaliana photosystem I(PSI-LHCII-ST2) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7A
 
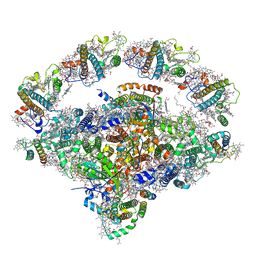 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 1 (PSI-ST1) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
7E1Q
 
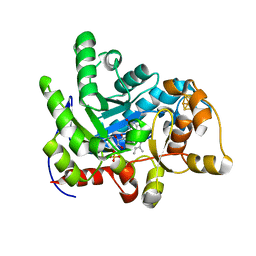 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1R
 
 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
2LDJ
 
 | | 1H Chemical Shift Assignments and structure of Trp-Cage mini-protein with D-amino acid | | Descriptor: | Trp-Cage mini-protein | | Authors: | Granillo, A.R, Annavarapu, S, Zhang, L, Koder, R, Nanda, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational Design of Thermostabilizing d-Amino Acid Substitutions.
J.Am.Chem.Soc., 133, 2011
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
6N7X
 
 | | S. cerevisiae U1 snRNP | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | Authors: | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
5YVD
 
 | |
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
4KFP
 
 | | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived Ureas as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[1-(tetrahydro-2H-pyran-4-yl)piperidin-4-yl]sulfonyl}benzyl)-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Bair, K.W, Baumeister, T, Ho, Y, Liederer, B.M, Liu, X, O'Brien, T, Oeh, J, Sampath, D, Skelton, N, Wang, L, Wang, W, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X. | | Deposit date: | 2013-04-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived ureas as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8OWZ
 
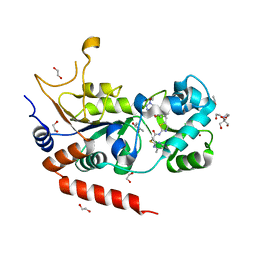 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
5B0J
 
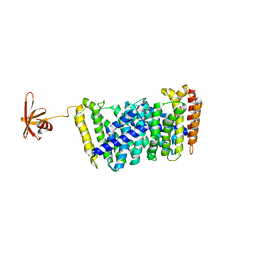 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
