6DO0
 
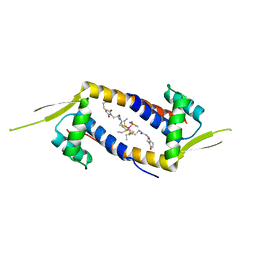 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex with Rhodium Bis-diphosphine Complex | | Descriptor: | Transcriptional regulator, PadR-like family, bis[diethyl(methyl)-lambda~5~-phosphanyl]{bis[{[(2-{[2-(2,5-dioxopyrrolidin-1-yl)ethyl]amino}-2-oxoethyl)amino]methyl}(diethyl)-lambda~5~-phosphanyl]}rhodium | | Authors: | Zadvornyy, O.A, Laureanti, J.A, Katipamula, S, O'Hagan, M, Peters, J.W. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Protein Scaffold Activates Catalytic CO2 Hydrogenation by a Rhodium Bis(diphosphine) Complex
Acs Catalysis, 9, 2019
|
|
5JFC
 
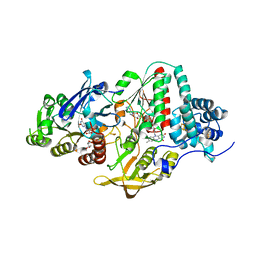 | | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, King, P.W, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
5JCA
 
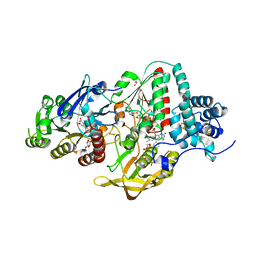 | | NADP(H) bound NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
6VWE
 
 | |
5VJ7
 
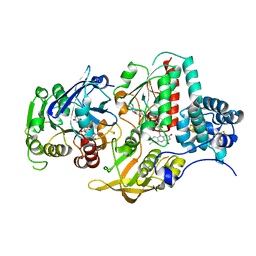 | | Ferredoxin NADP Oxidoreductase (Xfn) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP(+) reductase subunit alpha, ... | | Authors: | Zadvornyy, O.A, Nguyen, D.M.N, Schut, G.J, Lipscomb, G.L, Tokmina-Lukaszewska, M, Adams, M.W.W, Peters, J.W. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two functionally distinct NADP(+)-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus.
J. Biol. Chem., 292, 2017
|
|
6BBL
 
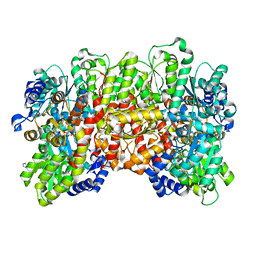 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
7MGN
 
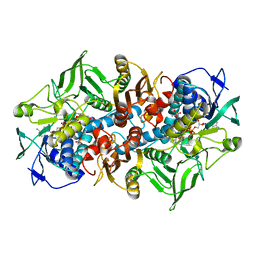 | | Crystal structure of F501H/H506E variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Zadvornyy, O.A, Prussia, G, Peters, J.W. | | Deposit date: | 2021-04-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
6N6P
 
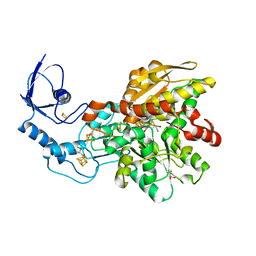 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6N59
 
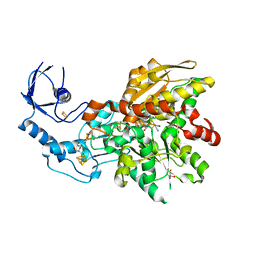 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
4Z1Y
 
 | |
4Z17
 
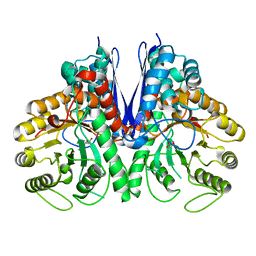 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
4YWS
 
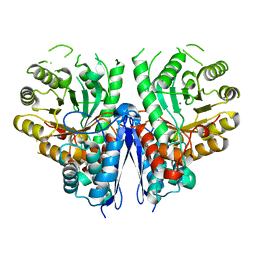 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
4XPI
 
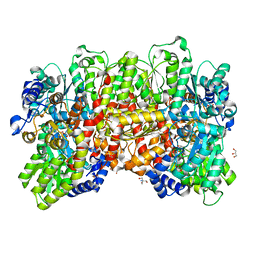 | | Fe protein independent substrate reduction by nitrogenase variants altered in intramolecular electron transfer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Danyal, K, Rasmusen, A.J, Keable, S.M, Shaw, S, Zadvornyy, O, Duval, S, Dean, D.R, Raugei, S, Peters, J.W, Seefeldt, L.C. | | Deposit date: | 2015-01-17 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fe protein-independent substrate reduction by nitrogenase MoFe protein variants.
Biochemistry, 54, 2015
|
|
6CDK
 
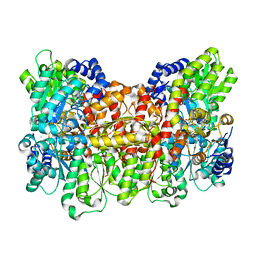 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
7MGO
 
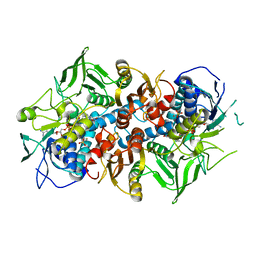 | | Crystal structure of F501H variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Prussia, G, Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
6NAC
 
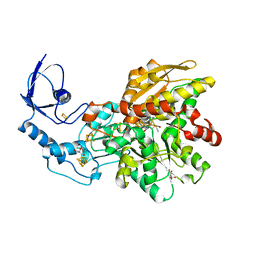 | | Crystal structure of [FeFe]-hydrogenase I (CpI) solved with single pulse free electron laser data | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Cohen, A.E, Davidson, C.M, Zadvornyy, O.A, Keable, S.M, Lyubimov, A.Y, Song, J, McPhillips, S.E, Soltis, S.M, Peters, J.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
4R0V
 
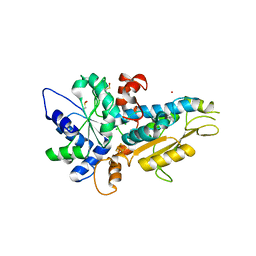 | | [FeFe]-hydrogenase Oxygen Inactivation is Initiated by the Modification and Degradation of the H cluster 2Fe Subcluster | | Descriptor: | ARSENIC, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Swanson, S.D, Ratzloff, M.W, Mulder, D.W, Artz, J.H, Ghose, S, Hoffman, A, White, S, Zadvornyy, O.A, Broderick, J.B, Bothner, B, King, P.W, Peters, J.W. | | Deposit date: | 2014-08-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster.
J.Am.Chem.Soc., 137, 2015
|
|
4YWO
 
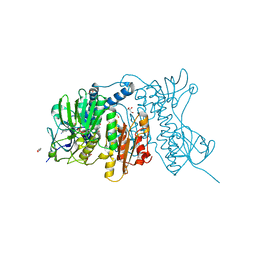 | | Mercuric reductase from Metallosphaera sedula | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase | | Authors: | Artz, J.H, Zadvornyy, O.A, White, S, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula.
Front Bioeng Biotechnol, 3, 2015
|
|
