4S3H
 
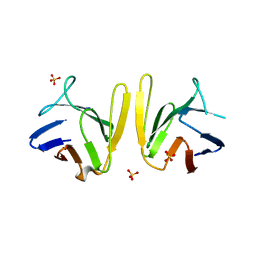 | | Crystal structure of S. pombe Mdb1 FHA domain | | Descriptor: | Mdb1, SULFATE ION | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1.
J.Biol.Chem., 290, 2015
|
|
5WY4
 
 | |
5WYL
 
 | |
5WY3
 
 | |
6KE6
 
 | |
6LQS
 
 | |
6LQR
 
 | |
5YDU
 
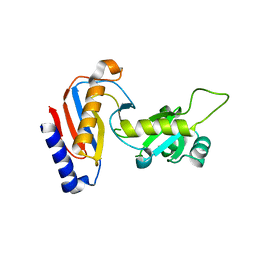 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
6LQV
 
 | |
6LQT
 
 | |
6LQQ
 
 | |
6LQP
 
 | |
6LQU
 
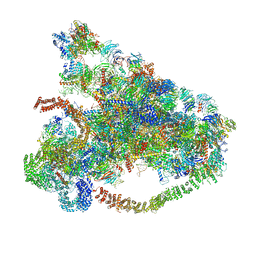 | |
6LZ1
 
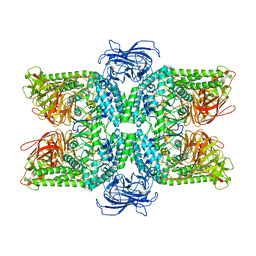 | | Structure of S.pombe alpha-mannosidase Ams1 | | Descriptor: | Ams1, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of fission yeast tetrameric alpha-mannosidase Ams1.
Febs Open Bio, 10, 2020
|
|
5YMA
 
 | | Crystal structure of ribosome assembly factor Efg1 | | Descriptor: | Putative rRNA processing protein | | Authors: | Shu, S, Ye, K. | | Deposit date: | 2017-10-21 | | Release date: | 2018-01-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Structural and functional analysis of ribosome assembly factor Efg1.
Nucleic Acids Res., 46, 2018
|
|
5YZ4
 
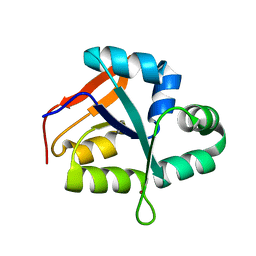 | | Structure of the PIN domain endonuclease Utp24 | | Descriptor: | CALCIUM ION, ZINC ION, rRNA-processing protein fcf1 | | Authors: | Du, Y, An, W, Ye, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.
Plos One, 13, 2018
|
|
5Z1G
 
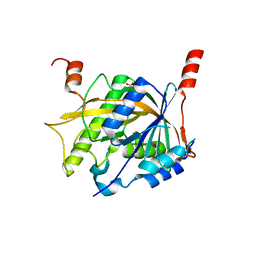 | | Structure of the Brx1 and Ebp2 complex | | Descriptor: | Ribosome biogenesis protein BRX1, SULFATE ION, rRNA-processing protein EBP2 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2017-12-26 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein Cell, 10, 2019
|
|
5Z3G
 
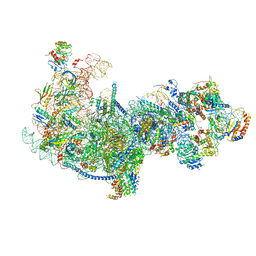 | | Cryo-EM structure of a nucleolar pre-60S ribosome (Rpf1-TAP) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Zhu, X, Zhou, D, Ye, K. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate.
Protein Cell, 10, 2019
|
|
8KHQ
 
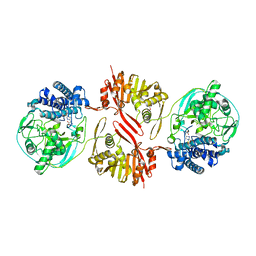 | | Bifunctional sulfoxide synthase OvoA_Th2 in complex with histidine and cysteine | | Descriptor: | 5-histidylcysteine sulfoxide synthase/putative 4-mercaptohistidine N1-methyltranferase, COBALT (II) ION, CYSTEINE, ... | | Authors: | Wang, J, Ye, K, Wang, X.Y, Yan, W.P. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Biochemical and Structural Characterization of OvoA Th2 : A Mononuclear Nonheme Iron Enzyme from Hydrogenimonas thermophila for Ovothiol Biosynthesis.
Acs Catalysis, 13, 2023
|
|
7XPL
 
 | |
7DDE
 
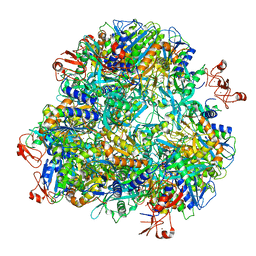 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7D63
 
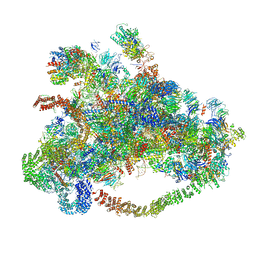 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5T
 
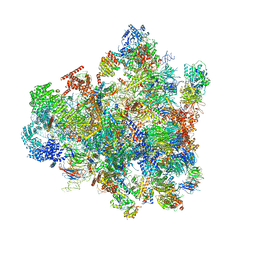 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
7D5S
 
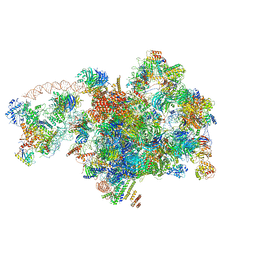 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
