1RS7
 
 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
4M3O
 
 | | Crystal structure of K.lactis Rtr1 NTD | | Descriptor: | KLLA0F12672p, ZINC ION | | Authors: | Hsu, P.L, Yang, W, Zheng, N, Varani, G. | | Deposit date: | 2013-08-06 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rtr1 Is a Dual Specificity Phosphatase That Dephosphorylates Tyr1 and Ser5 on the RNA Polymerase II CTD.
J.Mol.Biol., 426, 2014
|
|
4DNJ
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4BS1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DXY
 
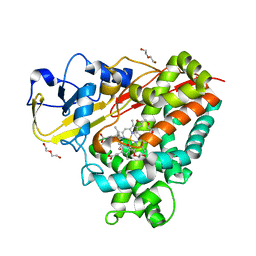 | | Crystal structures of CYP101D2 Y96A mutant | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Bell, S.G, Yang, W, Dale, A, Wong, L.-L. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the affinity and activity of CYP101D2 for hydrophobic substrates
Appl.Microbiol.Biotechnol., 2012
|
|
4E44
 
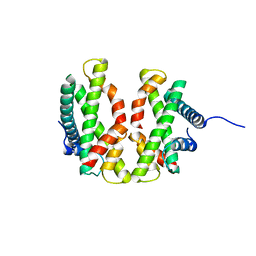 | |
6OZP
 
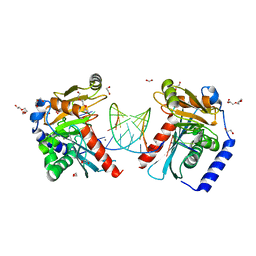 | |
6OZS
 
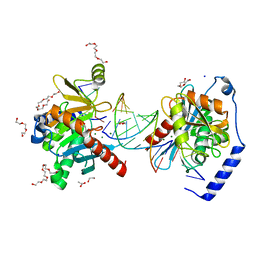 | |
6OZK
 
 | |
6OZR
 
 | |
6OZO
 
 | |
6OZN
 
 | |
5XGK
 
 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
3NGO
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
1ETK
 
 | |
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
5C8Z
 
 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | Descriptor: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1ETO
 
 | |
1EA6
 
 | |
1ETQ
 
 | |
1H7S
 
 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
2AXC
 
 | | Crystal structure of ColE7 translocation domain | | Descriptor: | Colicin E7, GLYCEROL, SULFATE ION | | Authors: | Cheng, Y.S, Shi, Z, Doudeva, L.G, Yang, W.Z, Chak, K.F, Yuan, H.S. | | Deposit date: | 2005-09-04 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes
J.Mol.Biol., 356, 2006
|
|
1H7U
 
 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1JX4
 
 | | Crystal Structure of a Y-family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*TP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-05 | | Release date: | 2001-10-05 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
