2LR7
 
 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
6M6P
 
 | | Structure of Marine bacterial laminarinase mutant E135A in complex with 1,3-beta-cellotriosyl-glucose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Tanokura, M, Long, L, Miyakawa, T. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6NRY
 
 | |
7VQM
 
 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
7VW1
 
 | |
7VW0
 
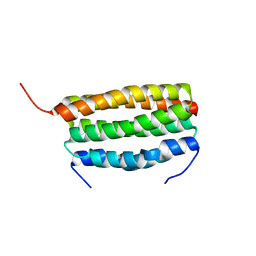 | | Structure of a dimeric periplasmic protein | | Descriptor: | DUF305 domain-containing protein | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle.
J.Inorg.Biochem., 229, 2022
|
|
7CBZ
 
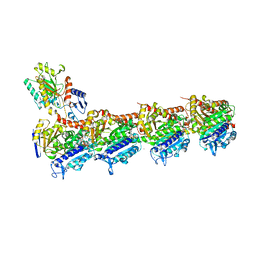 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
8HPG
 
 | |
7CUX
 
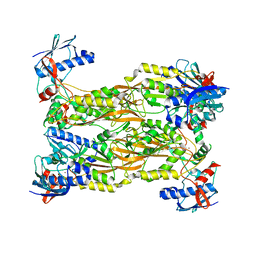 | | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Yang, J.Y, Luo, M, Ou, J.Y, Wang, Z.W, Gao, S. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29477072 Å) | | Cite: | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding
To Be Published
|
|
7D6N
 
 | |
7E5C
 
 | | Bacterial prolidase mutant D45W/L225Y/H226L/H343I | | Descriptor: | MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Yang, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis
Catalysis Science And Technology, 11, 2021
|
|
7EMJ
 
 | | Crystal structure of T2R-TTL-Barbigerone complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8,8-dimethyl-3-(2,4,5-trimethoxyphenyl)pyrano[2,3-f]chromen-4-one, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of T2R-TTL-barbigerone complex
To Be Published
|
|
7CCB
 
 | |
7D6M
 
 | |
7F0Y
 
 | |
7F13
 
 | | Crystal structure of isomerase Dcr3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dcr3 | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7F0Z
 
 | | Crystal structure of NsrQ W31A | | Descriptor: | NsrQ | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7F11
 
 | | Crystal structure of NsrQ M128I in complex with substrate analogue 7 | | Descriptor: | NsrQ, methyl 2-[2,6-bis(oxidanyl)phenyl]carbonyl-5-methyl-3,6-bis(oxidanyl)benzoate | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7F0O
 
 | |
7F10
 
 | |
7F14
 
 | |
7ECC
 
 | | M4 family peptidase PlM4P-mature form | | Descriptor: | CALCIUM ION, M4 family peptidase, PHOSPHATE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | M4 family peptidase PlM4P-mature form
To Be Published
|
|
7FGZ
 
 | | Marine bacterial GH16 hydrolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GH16 hydrolase | | Authors: | Yang, J. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Marine bacterial GH16 hydrolase
To Be Published
|
|
6TL0
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
6TKY
 
 | | Crystal structure of the DHR2 domain of DOCK10 in complex with CDC42 | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10, GLYCEROL | | Authors: | Barford, D, Fan, D, Cronin, N, Yang, J. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for CDC42 and RAC activation by the dual specificity GEF DOCK10
Biorxiv, 2022
|
|
