1Z8W
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192I) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8X
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192V) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8T
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192Q) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
3A6V
 
 | |
3A6T
 
 | | Crystal structure of MutT-8-OXO-DGMP complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, Mutator mutT protein, SODIUM ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3ACA
 
 | |
3A6U
 
 | | Crystal structure of MutT-8-OXO-dGMP-MN(II) complex | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mutator mutT protein, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3A6S
 
 | | Crystal structure of the MutT protein | | Descriptor: | L(+)-TARTARIC ACID, Mutator mutT protein, SODIUM ION | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2009-09-09 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base
J.Biol.Chem., 285, 2010
|
|
3AC9
 
 | |
5XDH
 
 | | His/DOPA ligated cytochrome c from an anammox organism KSU-1 | | Descriptor: | ACETATE ION, HEME C, Putative cytochrome c, ... | | Authors: | Hira, D, Kitamura, R, Nakamura, T, Yamagata, Y, Furukawa, K, Fujii, T. | | Deposit date: | 2017-03-28 | | Release date: | 2018-03-28 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Anammox Organism KSU-1 Expresses a Novel His/DOPA Ligated Cytochrome c.
J. Mol. Biol., 430, 2018
|
|
2Z3Q
 
 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
2Z3R
 
 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | GLYCEROL, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
3APV
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APW
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APU
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein | | Descriptor: | Alpha-1-acid glycoprotein 2, TETRAETHYLENE GLYCOL | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
7WWA
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 2.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW7
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW9
 
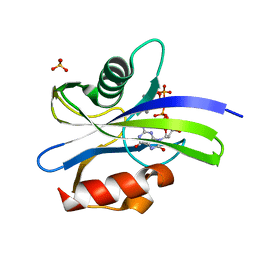 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW5
 
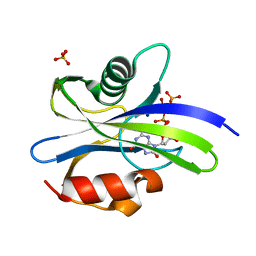 | | Crystal structure of MutT-8-oxo-dGTP complex | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW8
 
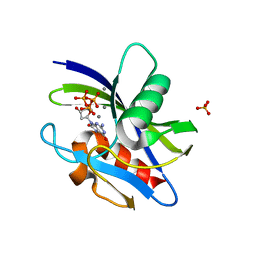 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 5 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW6
 
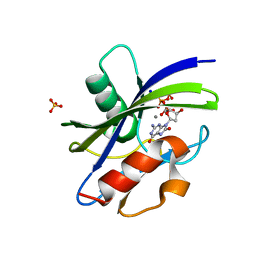 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 20 min in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X9L
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 4 hr using 10 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X9I
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 12 hr using 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X9K
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 2 hr using 10 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
