6NSB
 
 | |
6NSA
 
 | |
6NSG
 
 | |
6NSC
 
 | | Crystal structure of the A/Brisbane/10/2007 (H3N2) influenza virus hemagglutinin G186V/L194P mutant apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6NS9
 
 | | Crystal structure of the IVR-165 (H3N2) influenza virus hemagglutinin apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preventing an Antigenically Disruptive Mutation in Egg-Based H3N2 Seasonal Influenza Vaccines by Mutational Incompatibility.
Cell Host Microbe, 25, 2019
|
|
6NSF
 
 | |
3BJF
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Wu, N. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
3BJT
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | MAGNESIUM ION, OXALATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Wu, N. | | Deposit date: | 2007-12-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
1LOQ
 
 | |
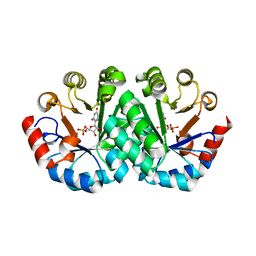
1LOS
 
 | |
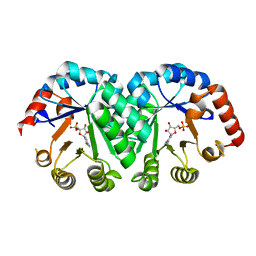
1LP6
 
 | |
1LOL
 
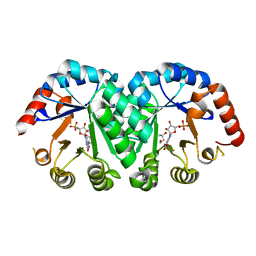 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
1LOR
 
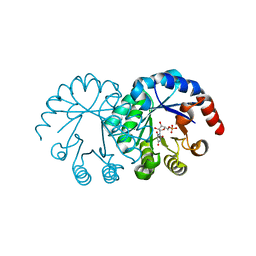 | | crystal structure of orotidine 5'-monophosphate complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, orotidine monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
6NHQ
 
 | |
6NHR
 
 | |
6NHP
 
 | |
7JN5
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 heavy chain, ... | | Authors: | Wu, N.C, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Plos Pathog., 16, 2020
|
|
7JMP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7JMO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
5VTR
 
 | |
5VU4
 
 | |
5VTW
 
 | |
5VTQ
 
 | |
5VTV
 
 | |
5VTY
 
 | |
