7ENM
 
 | |
7ENR
 
 | |
8HVR
 
 | |
8ITT
 
 | |
8ITR
 
 | |
8JKE
 
 | | AfsR(T337A) transcription activation complex | | Descriptor: | DNA(65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural and functional characterization of AfsR, an SARP family transcriptional activator of antibiotic biosynthesis in Streptomyces.
Plos Biol., 22, 2024
|
|
7K6B
 
 | |
5W87
 
 | |
5WYI
 
 | | The Yaf9 YEATS domain Recognizing H3K122suc Peptide | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, ILE-MET-PRO-LYS-ASP-ILE-GLN-LEU, SUCCINIC ACID, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Yaf9 YEATS domain Recognizing H3K122suc Peptide
To Be Published
|
|
5Y8V
 
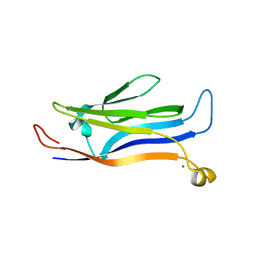 | | Crystal structure of GAS41 | | Descriptor: | MAGNESIUM ION, YEATS domain-containing protein 4 | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-08-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BLQ
 
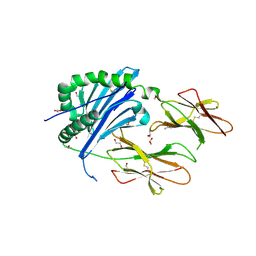 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BLX
 
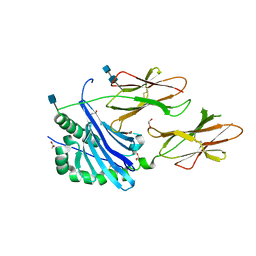 | | Crystal structure of IAg7 in complex with insulin mimotope p8G9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BLR
 
 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E6SS | | Descriptor: | 1,2-ETHANEDIOL, H2-Ab1 protein, IAg7 alpha chain | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7W5E
 
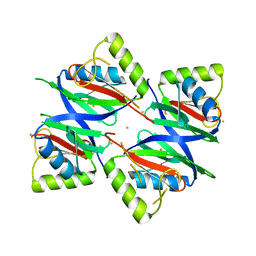 | | Oxidase ChaP D49L mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7WA8
 
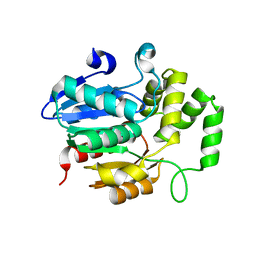 | | Strigolactone receptors in Striga ShHTL7 | | Descriptor: | Hyposensitive to light 7 | | Authors: | Wang, Y, Yao, R. | | Deposit date: | 2021-12-12 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga.
Plant Physiology, Volume 185, 2021
|
|
5XKF
 
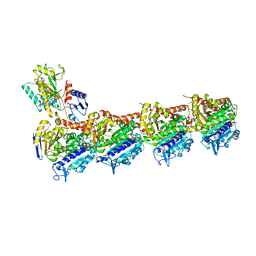 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKE
 
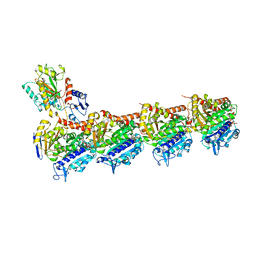 | | Crystal structure of T2R-TTL-Demecolcine complex | | Descriptor: | (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKG
 
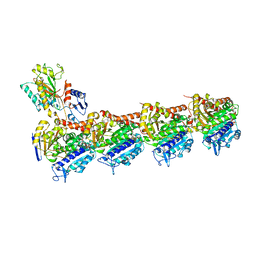 | | Crystal structure of T2R-TTL-CH1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(3-azanyl-4-methoxy-phenyl)-methyl-amino]chromen-2-one, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKH
 
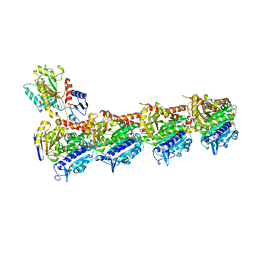 | | Crystal structure of T2R-TTL-CF1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(4-methoxy-3-oxidanyl-phenyl)-methyl-amino]chromen-2-one, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
4N74
 
 | |
4U7U
 
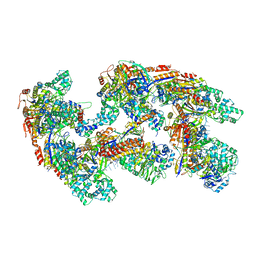 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
4W9O
 
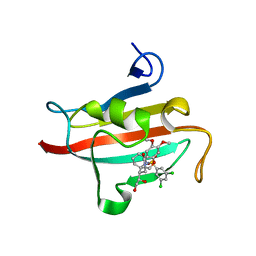 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1AHC
 
 | |
1AHB
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ALPHA-MOMORCHARIN, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1AHA
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ADENINE, ALPHA-MOMORCHARIN | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
