8JCZ
 
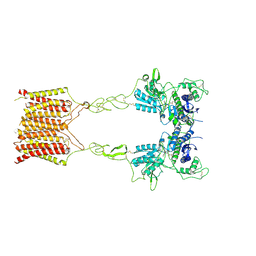 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
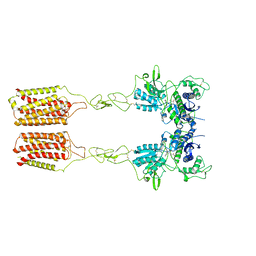 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD0
 
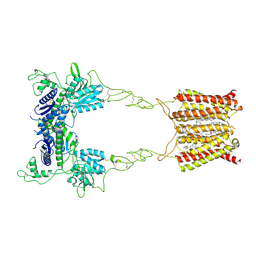 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
7WLC
 
 | |
7WED
 
 | |
7WEA
 
 | |
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEE
 
 | |
7WEC
 
 | |
7WEB
 
 | |
7WE9
 
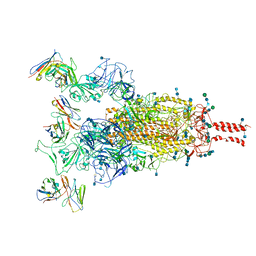 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEF
 
 | |
5ZUD
 
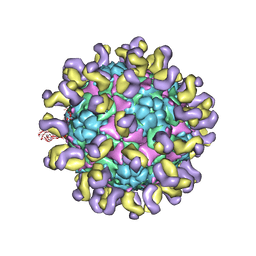 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
7X2H
 
 | |
6NJD
 
 | |
6NII
 
 | |
7XD2
 
 | |
5ZUF
 
 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
8GSE
 
 | | Echovirus3 capsid protein in complex with 6D10 Fab (upright) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSD
 
 | | Echovirus3 full particle in complex with 6D10 Fab | | Descriptor: | Genome polyprotein, Genome polyprotein (Fragment), Heavy chain of 6D10, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSF
 
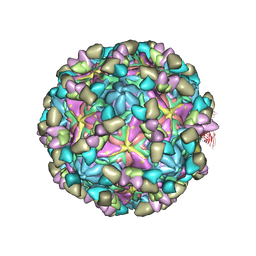 | | Echovirus3 empty particle in complex with 6D10 Fab (sideling) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSC
 
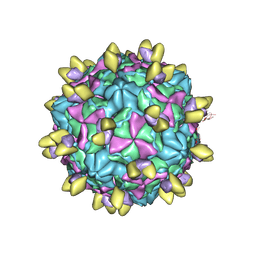 | | Echovirus3 A-particle in complex with 6D10 Fab | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP1, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
2JT8
 
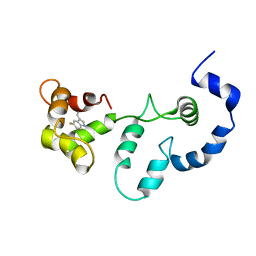 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2JT3
 
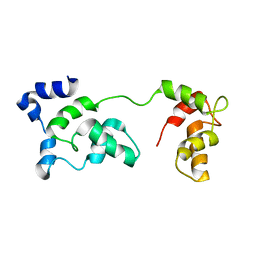 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
