6FLH
 
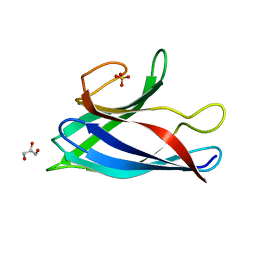 | | Monomeric Human Cu,Zn Superoxide dismutase, SOD1 7+7, apo form | | Descriptor: | GLYCEROL, SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Yang, F, Logan, D, Oliveberg, M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Cost of Long Catalytic Loops in Folding and Stability of the ALS-Associated Protein SOD1.
J.Am.Chem.Soc., 140, 2018
|
|
7U2K
 
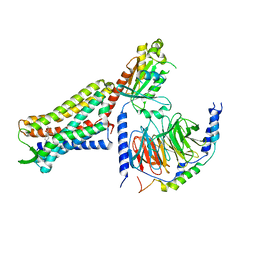 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
4I15
 
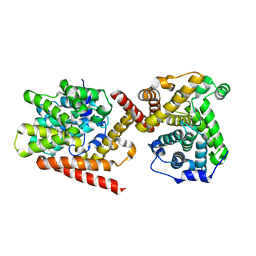 | | Crystal structure of TbrPDEB1 | | Descriptor: | Class 1 phosphodiesterase PDEB1, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel Trypanosoma brucei Phosphodiesterase B1 Inhibitors by Virtual Screening against the Unliganded TbrPDEB1 Crystal Structure.
J.Med.Chem., 56, 2013
|
|
3WOH
 
 | |
1BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
4ED5
 
 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QBS
 
 | |
8G9E
 
 | |
4QBR
 
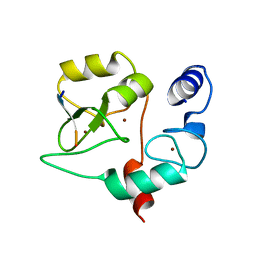 | |
8R0F
 
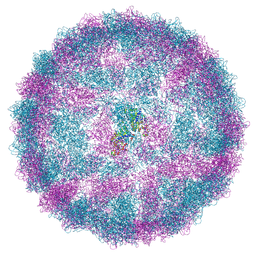 | | Capsid structure of Giardiavirus (GLV) HP strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) HP strain
To Be Published
|
|
4ZYO
 
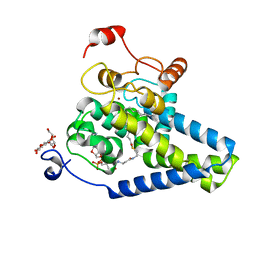 | | Crystal Structure of Human Integral Membrane Stearoyl-CoA Desaturase with Substrate | | Descriptor: | Acyl-CoA desaturase, DODECYL-BETA-D-MALTOSIDE, STEAROYL-COENZYME A, ... | | Authors: | Wang, H, Klein, M.G, Lane, W, Snell, G, Levin, I, Li, K, Zou, H, Sang, B.-C. | | Deposit date: | 2015-05-21 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8R0G
 
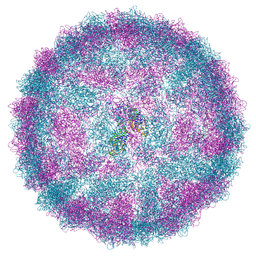 | | Capsid structure of Giardiavirus (GLV) CAT strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) CAT strain
To Be Published
|
|
3PFN
 
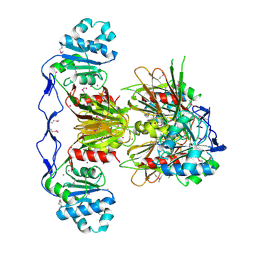 | | Crystal Structure of human NAD kinase | | Descriptor: | NAD kinase, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Tempel, W, Wernimont, A.K, Tong, Y, Guan, X, Shen, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of human NAD kinase
to be published
|
|
3Q3J
 
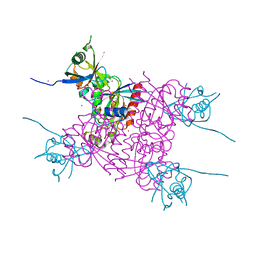 | | Crystal structure of plexin A2 RBD in complex with Rnd1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A2, ... | | Authors: | Wang, H, Tempel, W, Tong, Y, Guan, X, Shen, L, Buren, L, Zhang, N, Wernimont, A.K, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-21 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Crystal structure of plexin A2 RBD in complex with Rnd1
to be published
|
|
4RGW
 
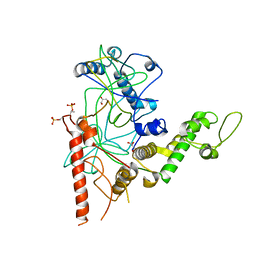 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
6HBA
 
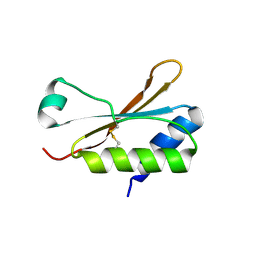 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBC
 
 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBB
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
4MMD
 
 | |
4MM4
 
 | |
4MM8
 
 | |
4MMF
 
 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM9
 
 | |
4MM5
 
 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMB
 
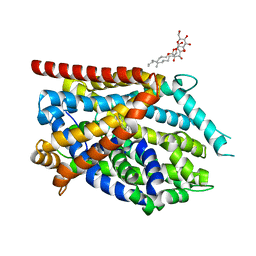 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
