3UEI
 
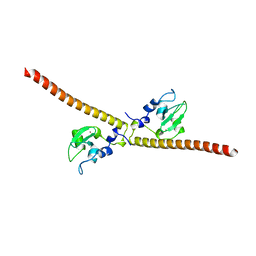 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEM
 
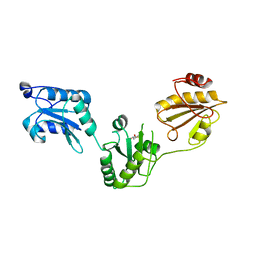 | | Crystal structure of human PDI bb'a' domains | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Protein disulfide-isomerase | | Authors: | Yu, J, Wang, C, Huo, L, Feng, W, Wang, C.-C. | | Deposit date: | 2011-10-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a'
J.Biol.Chem., 287, 2012
|
|
3UDO
 
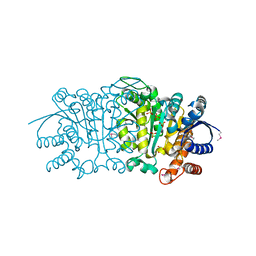 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
3UEF
 
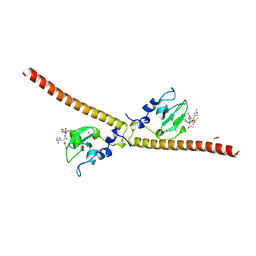 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UI4
 
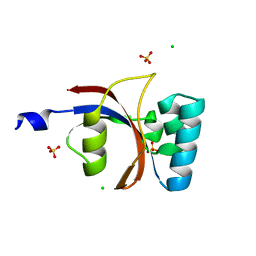 | | 0.8 A resolution crystal structure of human Parvulin 14 | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SULFATE ION | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
3UNE
 
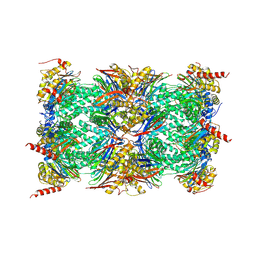 | | Mouse constitutive 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3ULR
 
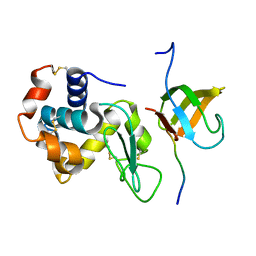 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | Descriptor: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | Authors: | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2011-11-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7QKC
 
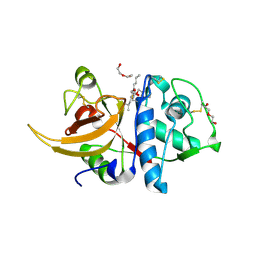 | | Crystal structure of human Cathepsin L after incubation with Sulfo-Calpeptin | | Descriptor: | Calpeptin, Cathepsin L, DI(HYDROXYETHYL)ETHER | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKA
 
 | | Crystal structure of SARS-CoV-2 Main Protease in complex with covalently bound GC376 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
5WZ3
 
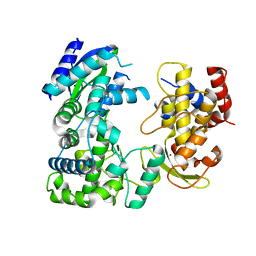 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase(RdRP) | | Descriptor: | NS5 RdRp, ZINC ION | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
4ESJ
 
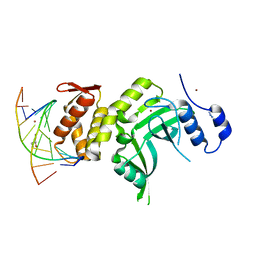 | | RESTRICTION ENDONUCLEASE DpnI IN COMPLEX WITH TARGET DNA | | Descriptor: | AZIDE ION, DNA (5'-D(*CP*TP*GP*GP*(6MA)P*TP*CP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Siwek, W, Czapinska, H, Bochtler, M, Bujnicki, J.M, Skowronek, K. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI.
Nucleic Acids Res., 40, 2012
|
|
4ETE
 
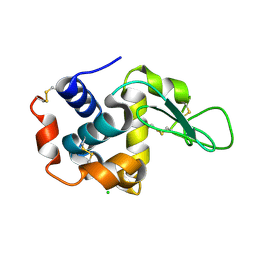 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4EUY
 
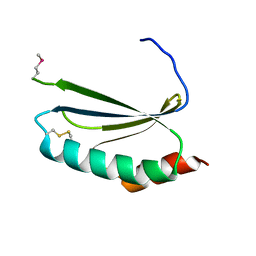 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
5X29
 
 | |
5WPY
 
 | |
5W96
 
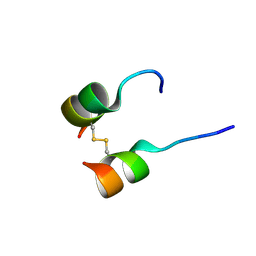 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5WPX
 
 | |
4EYY
 
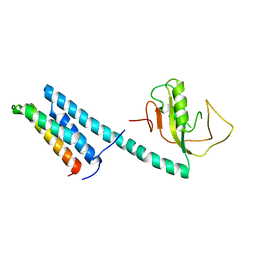 | | Crystal Structure of the IcmR-IcmQ complex from Legionella pneumophila | | Descriptor: | IcmQ, IcmR | | Authors: | Farelli, J.D, Gumbart, J, Akey, I.V, Amyot, W, Hempstead, A.D, Head, J.F, McKnight, C.J, Isberg, R.R, Akey, C.W. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | IcmQ in the Type 4b secretion system contains an NAD+ binding domain.
Structure, 21, 2013
|
|
5X41
 
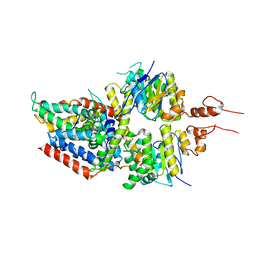 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
4EZA
 
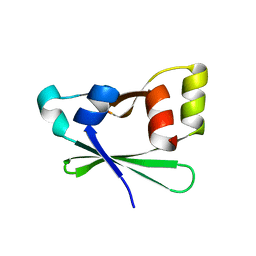 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
4F13
 
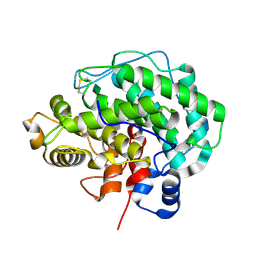 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5X93
 
 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-03-05 | | Release date: | 2017-08-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5X9O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
4FC2
 
 | |
