6WS2
 
 | |
6WS4
 
 | | Crystal structure of KRAS-G12D/K104Q mutant, GDP-bound | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T.H, Simanshu, D.K. | | Deposit date: | 2020-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Allosteric regulation of switch-II domain controls KRAS oncogenicity.
Cancer Res., 2023
|
|
6XHA
 
 | | Crystal Structure of KRAS-G12V (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XGV
 
 | | Crystal Structure of KRAS-G13D (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XHB
 
 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form II) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XGU
 
 | | Crystal Structure of KRAS-Q61R (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XCO
 
 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Jones, M.C, Tresoldi, E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6XC9
 
 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Tresoldi, M.C.E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6XCP
 
 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Jones, M.C, Tresoldi, E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
6VC8
 
 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TY1
 
 | | Crystal structure of apo eosinophil cationic protein (ribonuclease 3) from Macaca fascicularis (MfECP) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Eosinophil cationic protein, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8G9A
 
 | | Crystal structure of a resurrected ancestor (AncRNase) of the pancreatic-type RNases 2 and 3 sub-families | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2023-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8F5X
 
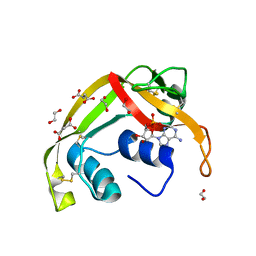 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8T73
 
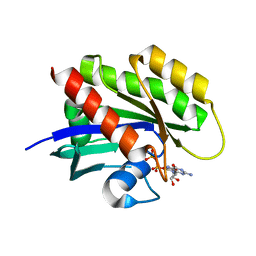 | | Crystal structure of KRAS4a-R151G with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T72
 
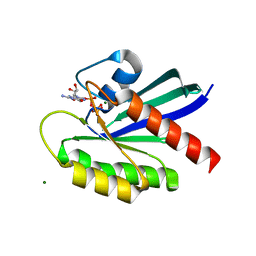 | | Crystal structure of WT KRAS4a with bound GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T71
 
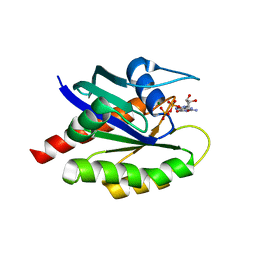 | | Crystal Structure of WT KRAS4a with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
6VJJ
 
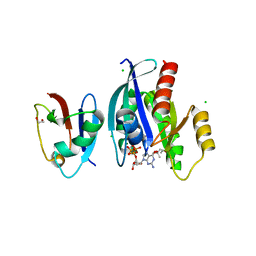 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6P4Z
 
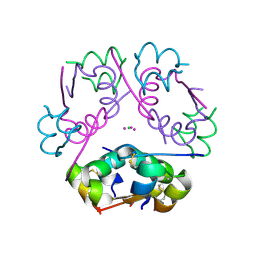 | | Structure of gadolinium-caged cobalt (III) insulin hexamer | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GADOLINIUM ATOM, ... | | Authors: | Tran, T.H, Taylor, S.K, Stojanovic, M.N, Tong, L. | | Deposit date: | 2019-05-29 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insulin Hexamer-Caged Gadolinium Ion as MRI Contrast-o-phore
ChemBioChem, 24, 2018
|
|
6OB3
 
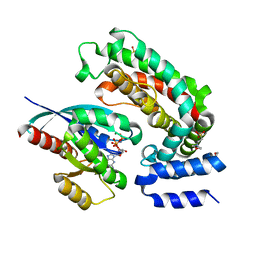 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB2
 
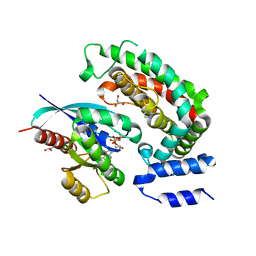 | | Crystal structure of wild-type KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8EPW
 
 | | Crystal Structure of KRAS4b-G13D (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2022-10-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reduced dynamic complexity allows structure elucidation of an excited state of KRAS G13D .
Commun Biol, 6, 2023
|
|
5GK5
 
 | | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution
To Be Published
|
|
8STN
 
 | | Crystal structure of KRAS-G12D/G75A mutant, GDP-bound | | Descriptor: | CHLORIDE ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
8STM
 
 | | Crystal structure of KRAS-G75A mutant, GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
5GK3
 
 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution
To Be Published
|
|
