8DD6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant in Complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, DIMETHYL SULFOXIDE, ORF1a polyprotein | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-17 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8DDL
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8SG6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Reduced with 20mM TCEP | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Tran, N, McLeod, M.J, Barwell, S, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
2JVC
 
 | |
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI2
 
 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
6Y93
 
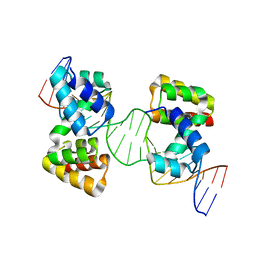 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
7NG0
 
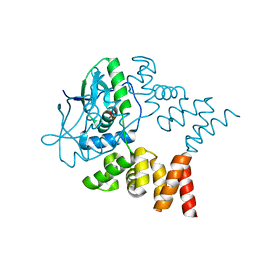 | | Crystal structure of N- and C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
7NFU
 
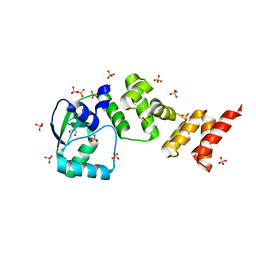 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
8SBR
 
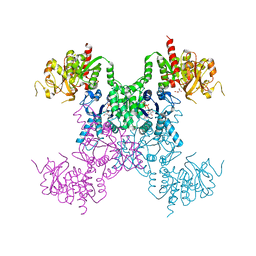 | |
6F5Z
 
 | |
6H2V
 
 | | Crystal structure of human METTL5-TRMT112 complex, the 18S rRNA m6A1832 methyltransferase at 2.5A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | van Tran, N, Graille, M. | | Deposit date: | 2018-07-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112.
Nucleic Acids Res., 47, 2019
|
|
6H2U
 
 | | Crystal structure of human METTL5-TRMT112 complex, the 18S rRNA m6A1832 methyltransferase at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, Methyltransferase-like protein 5, Multifunctional methyltransferase subunit TRM112-like protein, ... | | Authors: | van Tran, N, Graille, M. | | Deposit date: | 2018-07-16 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112.
Nucleic Acids Res., 47, 2019
|
|
8FV9
 
 | | E coli. CTP synthase in complex with F-dCTP | | Descriptor: | 2'-deoxy-2'-fluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVE
 
 | | E coli. CTP synthase in complex with CTP (potassium malonate + 100 mM MgCl2) | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV6
 
 | | E coli. CTP synthase in complex with dF-dCTP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV8
 
 | | E coli. CTP synthase in complex with dF-dCTP + ADP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVD
 
 | | E coli. CTP synthase in complex with dF-dCTP (potassium malonate + 100 mM MgCl2) | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MAGNESIUM ION, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVC
 
 | | E coli. CTP synthase in complex with dF-dCTP (potassium malonate + 5 mM MgCl2) | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV7
 
 | | E coli. CTP synthase in complex with dF-dCTP + ATP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVB
 
 | | E coli. CTP synthase in complex with CTP | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVA
 
 | | E coli. CTP synthase in complex with F-araCTP | | Descriptor: | 4-amino-1-{2-deoxy-2-fluoro-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, CTP synthase, MALONIC ACID, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
6S6H
 
 | | Crystal structure of the DNA binding domain of the chromosome-partitioning protein ParB complexed to the centromeric parS site | | Descriptor: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*C)-3'), GLYCEROL | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Tan, E.X, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2019-07-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
