6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
6RLN
 
 | | Crystal structure of RIP1 kinase in complex with GSK3145095 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[(3~{S})-7,9-bis(fluoranyl)-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepin-3-yl]-3-(phenylmethyl)-1~{H}-1,2,4-triazole-5-carboxamide | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer.
Acs Med.Chem.Lett., 10, 2019
|
|
6OCQ
 
 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | Descriptor: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
452D
 
 | | ACRIDINE BINDING TO DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Thorpe, J.H, Todd, A.K, Cardin, C.J. | | Deposit date: | 1999-02-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
J.Med.Chem., 42, 1999
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
6SKB
 
 | | Crystal Structure of Human Kallikrein 6 (N217D/I218Y/K224R) in complex with GSK3496783A | | Descriptor: | 4-[(3~{R})-1-oxidanyl-3,4-dihydro-2,1-benzoxaborinin-3-yl]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, 4-[(3~{S})-1-oxidanyl-3,4-dihydro-2,1-benzoxaborinin-3-yl]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design and development of a series of borocycles as selective, covalent kallikrein 5 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6SZG
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GR839 and GSK513 | | Descriptor: | (4-chlorophenyl)-[(3~{S})-3-oxidanylpiperidin-1-yl]methanone, 4,5,6,7-tetrahydro-2~{H}-indazole-3-carboxylic acid, CALCIUM ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6SKD
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK3397892A | | Descriptor: | 4-[[(3~{S})-1-oxidanyl-3,4-dihydro-2,1-benzoxaborinin-3-yl]methylamino]benzenecarboximidamide, GLYCEROL, Kallikrein-6, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and development of a series of borocycles as selective, covalent kallikrein 5 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6SKC
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK3448330A | | Descriptor: | 4-[(3~{S})-1-oxidanyl-3,4-dihydro-2,1-benzoxaborinin-3-yl]benzenecarboximidamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and development of a series of borocycles as selective, covalent kallikrein 5 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6SZH
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6QFG
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK144 | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QFE
 
 | | Crystal Structure of Human Kallikrein 5 in complex with GSK144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QFF
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK144 | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QHC
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK358180B | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Kallikrein-6, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHB
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK578724A | | Descriptor: | GLYCEROL, Kallikrein-6, ~{N}-(4-carbamimidoylphenyl)-3-methoxy-2-oxidanyl-benzamide | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHA
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3205388B | | Descriptor: | GLYCEROL, Kallikrein-6, UNKNOWN ATOM OR ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QFH
 
 | | Crystal Structure of Human Kallikrein 6 (N217D/I218Y/K224R) in complex with GSK144. | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QH9
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3239861A | | Descriptor: | (3~{R})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, (3~{S})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7A12
 
 | |
7A16
 
 | |
7A14
 
 | |
7A15
 
 | |
7A13
 
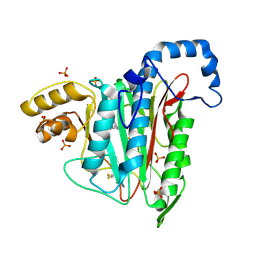 | |
1L4J
 
 | | Holliday Junction TCGGTACCGA with Na and Ca Binding Sites. | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', CALCIUM ION, SODIUM ION | | Authors: | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | Deposit date: | 2002-03-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational and Hydration Effects of Site-selective Sodium, Calcium and
Strontium Ion Binding to the DNA Holliday Junction Structure
d(TCGGTACCGA)4
J.Mol.Biol., 327, 2003
|
|
1JUC
 
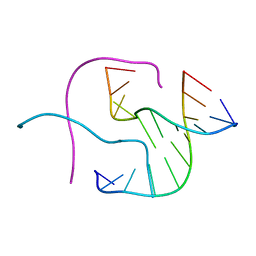 | | Crystal Structure Analysis of a Holliday Junction Formed by CCGGTACCGG | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3' | | Authors: | Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Cardin, C.J. | | Deposit date: | 2001-08-24 | | Release date: | 2002-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of a new crystal form of the four-way Holliday junction formed by the DNA sequence d(CCGGTACCGG)2: sequence versus lattice?
Acta Crystallogr.,Sect.D, 58, 2002
|
|
