2VO0
 
 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 1-[4-(4-chlorophenyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2X39
 
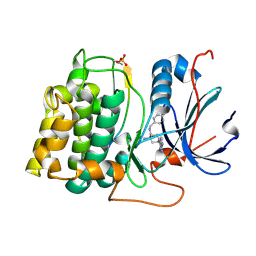 | | Structure of 4-Amino-N-(4-chlorobenzyl)-1-(7H-pyrrolo(2,3-d)pyrimidin- 4-yl)piperidine-4-carboxamide bound to PKB | | Descriptor: | 4-AMINO-N-(4-CHLOROBENZYL)-1-(7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PIPERIDINE-4-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, RAC-BETA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Davies, T.G, McHardy, T, Caldwell, J.J, Cheung, K.M, Hunter, L.J, Taylor, K, Rowlands, M, Ruddle, R, Henley, A, Brandon, A.D, Valenti, M, Fazal, L, Seavers, L, Raynaud, F.I, Eccles, S.A, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of 4-Amino-1-(7H-Pyrrolo[2,3-D]Pyrimidin-4-Yl)Piperidine-4-Carboxamides as Selective, Orally Active Inhibitors of Protein Kinase B (Akt).
J.Med.Chem., 53, 2010
|
|
5CHX
 
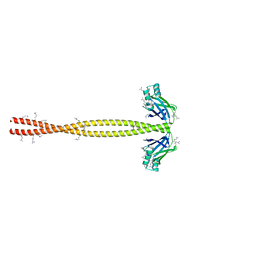 | | Crystal Structure of amino acids 1590-1657 of MYH7 | | Descriptor: | Xrcc4-MYH7-1590-1657 | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinz, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-10 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
5CJ0
 
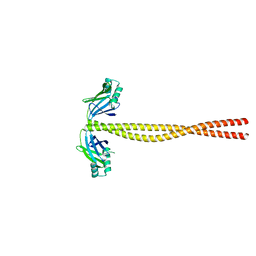 | | Crystal Structure of Amino Acids 1631-1692 of MYH7 | | Descriptor: | Xrcc4-MYH7-(1631-1692) chimera protein | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinze, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
5CJ4
 
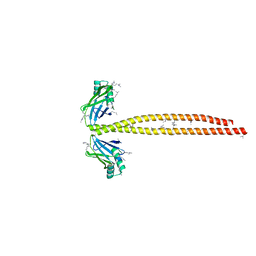 | | Crystal Structure of Amino Acids 1562-1622 of MYH7 | | Descriptor: | Xrcc4-MYH7-(1562-1622) chimera protein | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinze, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
1JXZ
 
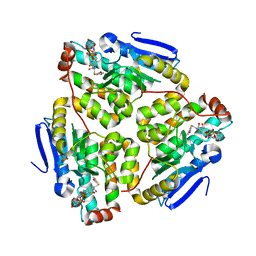 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
1N7P
 
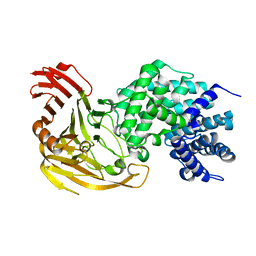 | | Streptococcus pneumoniae Hyaluronate Lyase W292A/F343V Double Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7Q
 
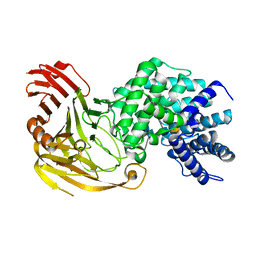 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A Double Mutant complex with hyaluronan hexasacchride | | Descriptor: | HYALURONIDASE, beta-D-galactopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7N
 
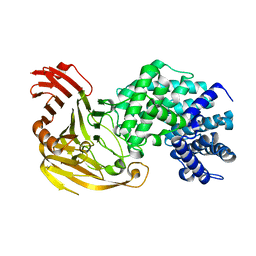 | | Streptococcus pneumoniae Hyaluronate Lyase W292A Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7O
 
 | | Streptococcus pneumoniae Hyaluronate Lyase F343V Mutant | | Descriptor: | hyaluronidase | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7R
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A/F343V Mutant complex with hexasaccharide hyaluronan | | Descriptor: | HYALURONIDASE, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
4KQK
 
 | | Crystal structure of CobT S80Y/Q88M/L175M complexed with p-cresol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQF
 
 | | Crystal structure of CobT E174A complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQH
 
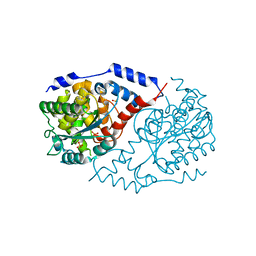 | | Crystal structure of CobT E317A | | Descriptor: | 1,2-ETHANEDIOL, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, PHOSPHATE ION, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQJ
 
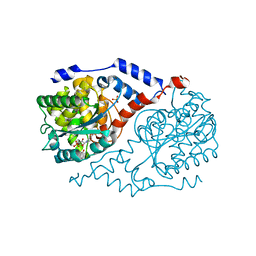 | | Crystal structure of CobT S80Y/Q88M/L175M complexed with p-cresol and NaMN | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, P-CRESOL, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQG
 
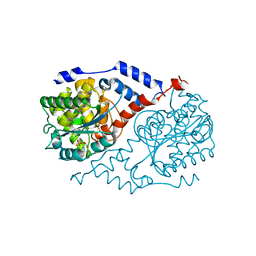 | | Crystal structure of CobT E174A complexed with DMB | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQI
 
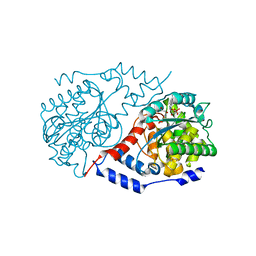 | | Crystal structure of CobT E317A complexed with its reaction products | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-RIBAZOLE-5'-PHOSPHATE, NICOTINIC ACID, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
