7RSI
 
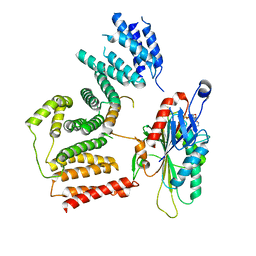 | | The cryo-EM map of KIF18A bound to KIFBP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF18A, ... | | Authors: | Tan, Z, Solon, A.L, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7RYQ
 
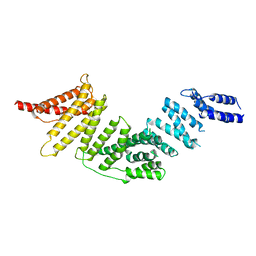 | | Cryo-EM map of KIFBP | | Descriptor: | KIF-binding protein | | Authors: | Tan, Z, Solon, A.L, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
5X38
 
 | |
5X35
 
 | |
5X34
 
 | |
5X39
 
 | |
5X37
 
 | |
7UBO
 
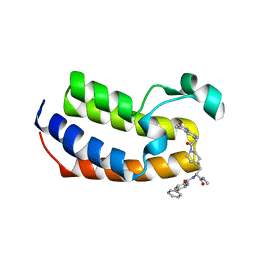 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor CCD-956 | | Descriptor: | Bromodomain testis-specific protein, DIMETHYL SULFOXIDE, N-[(2R)-1-(methylamino)-3-{1-[(4-methyl-2-oxo-1,2-dihydroquinolin-6-yl)acetyl]piperidin-4-yl}-1-oxopropan-2-yl]-5-phenylpyridine-2-carboxamide | | Authors: | Ta, H.M, Modukuri, R.K, Yu, Z, Tan, Z, Matzuk, M.M, Kim, C. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RSQ
 
 | | Cryo-EM structure of KIFBP core | | Descriptor: | KIF-binding protein | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7RYP
 
 | | Cryo-EM structure of KIFBP:KIF15 | | Descriptor: | KIF-binding protein, Kinesin-like protein KIF15 | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
5X3C
 
 | |
5X36
 
 | |
3UUO
 
 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2MWK
 
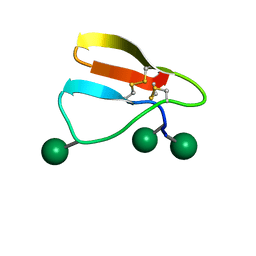 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWJ
 
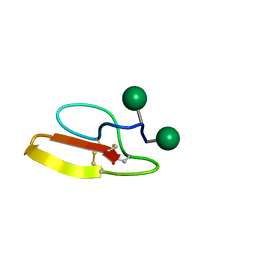 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
7YHG
 
 | |
7YHH
 
 | |
7YHF
 
 | |
7YHI
 
 | |
