2MEA
 
 | |
2MEB
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEF
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-04 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MED
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEI
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEH
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
4OEC
 
 | | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 | | Descriptor: | Glycerophosphoryl diester phosphodiesterase, MAGNESIUM ION | | Authors: | Atsuta, Y, You, D.J, Takano, K, Koga, Y, Kanaya, S. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1
To be Published
|
|
3U3G
 
 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
5NEN
 
 | |
7XNC
 
 | | MEK1 bound to DS94070624 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Kishikawa, S, Takano, K, Ubukata, O, Hanzawa, H. | | Deposit date: | 2022-04-28 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel ATP-Competitive MEK Inhibitor DS03090629 that Overcomes Resistance Conferred by BRAF Overexpression in BRAF-Mutated Melanoma.
Mol.Cancer Ther., 22, 2023
|
|
7XLP
 
 | | MEK1 bound to DS03090629 | | Descriptor: | (1~{R},3~{S})-3-[[6-[2-chloranyl-4-(4-methylpyrimidin-2-yl)oxy-phenyl]-3-methyl-1~{H}-indazol-4-yl]oxy]cyclohexan-1-amine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kishikawa, S, Takano, K, Ubukata, O, Hanzawa, H. | | Deposit date: | 2022-04-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel ATP-Competitive MEK Inhibitor DS03090629 that Overcomes Resistance Conferred by BRAF Overexpression in BRAF-Mutated Melanoma.
Mol.Cancer Ther., 22, 2023
|
|
6M4E
 
 | | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M55
 
 | | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M4F
 
 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
5AVH
 
 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVN
 
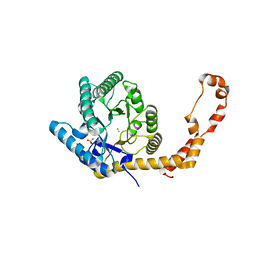 | | The 1.03 angstrom structure (P212121) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVD
 
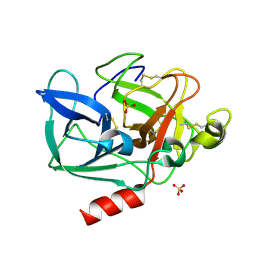 | | The 0.86 angstrom structure of elastase crystallized in high-strength agarose hydrogel | | Descriptor: | Chymotrypsin-like elastase family member 1, SULFATE ION | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVG
 
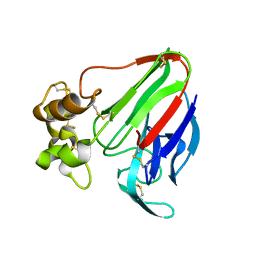 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5X7K
 
 | |
1C7P
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME WITH FOUR EXTRA RESIDUES (EAEA) AT THE N-TERMINAL | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Katakura, Y, Yutani, K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris.
Protein Eng., 13, 2000
|
|
3WR7
 
 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
2E1P
 
 | | Crystal structure of pro-Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Saito, K, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of unautoprocessed precursor of subtilisin from a hyperthermophilic archaeon: evidence for Ca2+-induced folding
J.Biol.Chem., 282, 2007
|
|
1YAO
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAP
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAM
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
