3WVS
 
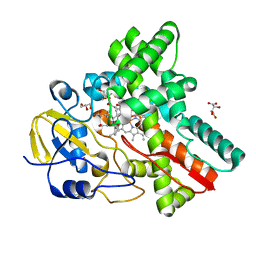 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
2E89
 
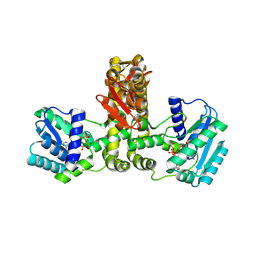 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ5
 
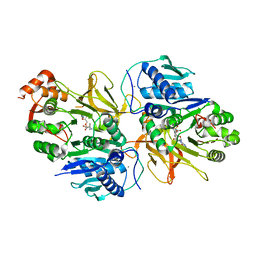 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
5YBW
 
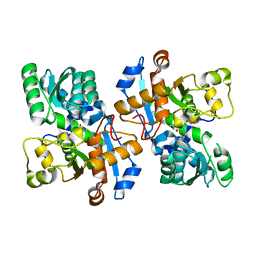 | | Crystal structure of pyridoxal 5'-phosphate-dependent aspartate racemase | | Descriptor: | Aspartate racemase | | Authors: | Mizobuchi, T, Nonaka, R, Yoshimura, M, Abe, K, Takahashi, S, Kera, Y, Goto, M. | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a pyridoxal 5'-phosphate-dependent aspartate racemase derived from the bivalve mollusc Scapharca broughtonii
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
