1P6T
 
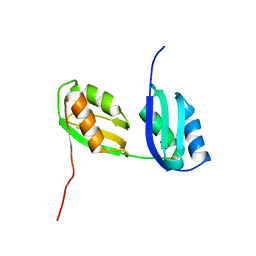 | | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis.
J.Biol.Chem., 278, 2003
|
|
1OQ3
 
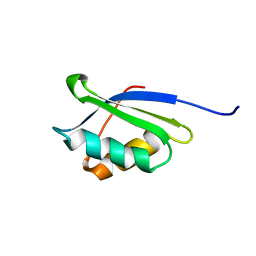 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1OPZ
 
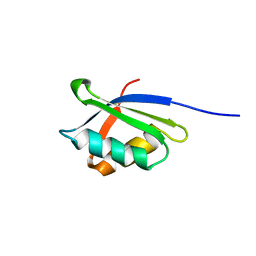 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonneli, L, Su, X.C. | | Deposit date: | 2003-03-06 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis.
J.Mol.Biol., 331, 2003
|
|
4E2A
 
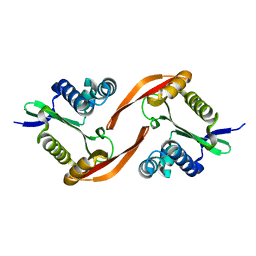 | |
1J6Q
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
3D3F
 
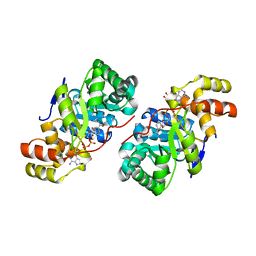 | |
4H10
 
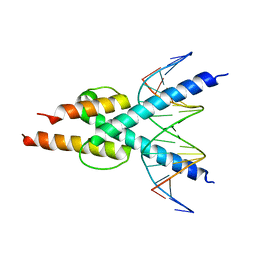 | |
2BB0
 
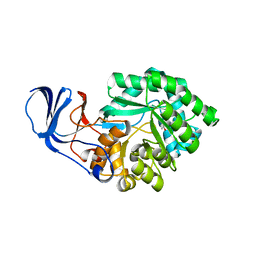 | |
2HVW
 
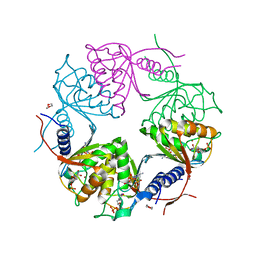 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
7EMK
 
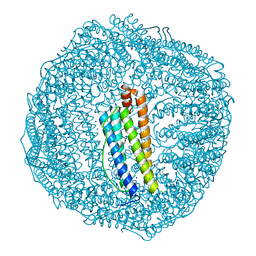 | | Dendrorhynchus zhejiangensis ferritin | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dendrorhynchus zhejiangensis ferritin, ... | | Authors: | Huan, H.S, Ming, T.H, Su, X.R. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-19 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of ferritin from Dendrorhynchus zhejiangensis
Biochemical and Biophysical Research Communications, 531, 2020
|
|
4F5D
 
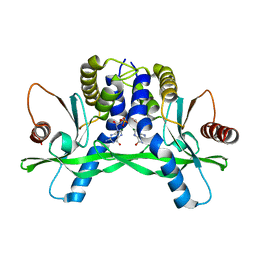 | | ERIS/STING in complex with ligand | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
4F5E
 
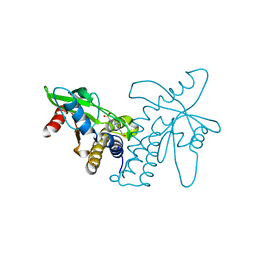 | | Crystal structure of ERIS/STING | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
6IJO
 
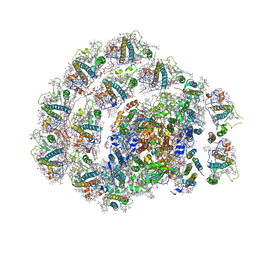 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
2G0J
 
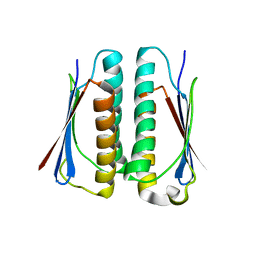 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
2B78
 
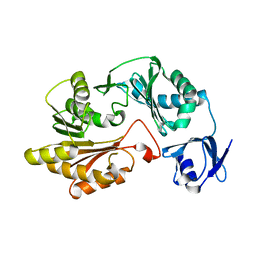 | |
2BAZ
 
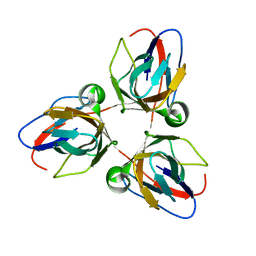 | |
2HVV
 
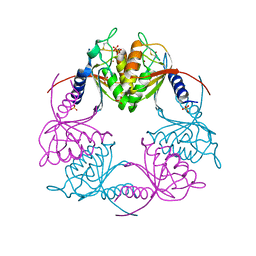 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | SULFATE ION, ZINC ION, deoxycytidylate deaminase | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
4IYR
 
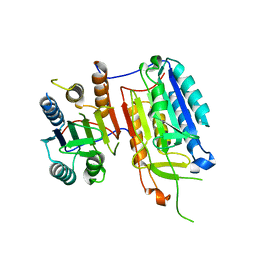 | | Crystal structure of full-length caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
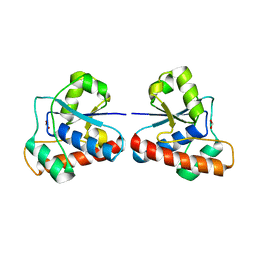
4EGS
 
 | |
2ZBB
 
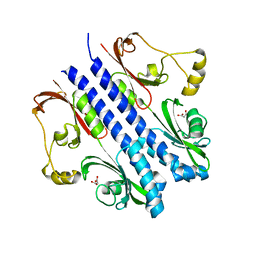 | | P43 crystal of DctBp | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID | | Authors: | Zhou, Y.F, Nan, B.Y, Liu, X, Nan, J, Liang, Y.H, Panjikar, S, Ma, Q.J, Wang, Y.P, Su, X.-D. | | Deposit date: | 2007-10-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | crystal structures of sensory histidine kinase DctBp
to be published
|
|
5SY5
 
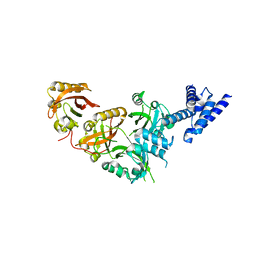 | | Crystal Structure of the Heterodimeric NPAS1-ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Neuronal PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
