3GFR
 
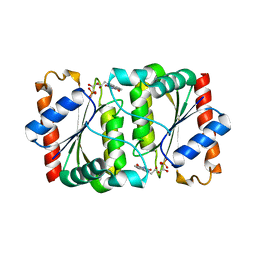 | | Structure of YhdA, D137L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3GFQ
 
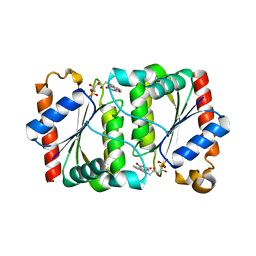 | | Structure of YhdA, K109L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3GFS
 
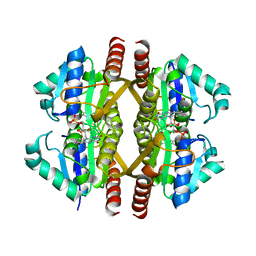 | | Structure of YhdA, K109D/D137K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3DQZ
 
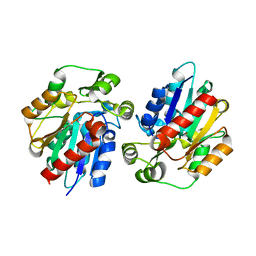 | | Structure of the hydroxynitrile lyase from Arabidopsis thaliana | | Descriptor: | Alpha-hydroxynitrile lyase-like protein, CHLORIDE ION | | Authors: | Andexer, J, Staunig, N, Gruber, K. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Hydroxynitrile lyases with alpha / beta-hydrolase fold: two enzymes with almost identical 3D structures but opposite enantioselectivities and different reaction mechanisms
Chembiochem, 13, 2012
|
|
