1CVM
 
 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1GS3
 
 | |
7D24
 
 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D26
 
 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | Descriptor: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D22
 
 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D25
 
 | | Hsp90 alpha N-terminal domain in complex with a 14 compund | | Descriptor: | 6-chloranyl-9-[(4-methylphenyl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D1V
 
 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | Descriptor: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
1H6L
 
 | |
1O9N
 
 | |
1OBI
 
 | |
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1O9O
 
 | |
1O9Q
 
 | |
1OCH
 
 | |
1OCK
 
 | |
1OBJ
 
 | |
1O9P
 
 | |
1OCL
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
 | |
1QLG
 
 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
1V0D
 
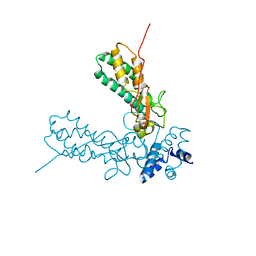 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
5IB0
 
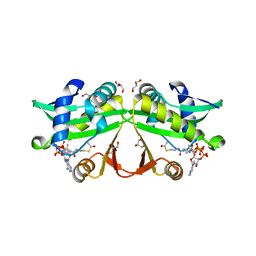 | | PA4534: acetyl CoA complex | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Uncharacterized protein PA4534 | | Authors: | Choe, J, Shin, S. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of PA4534
To Be Published
|
|
8ITN
 
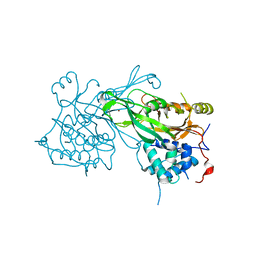 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design.
Commun Biol, 6, 2023
|
|
8ITP
 
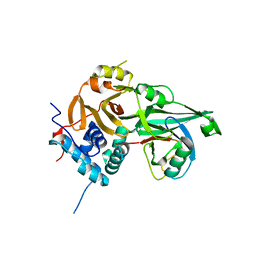 | |
