1J75
 
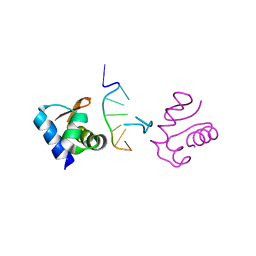 | | Crystal Structure of the DNA-Binding Domain Zalpha of DLM-1 Bound to Z-DNA | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Tumor Stroma and Activated Macrophage Protein DLM-1 | | Authors: | Schwartz, T, Behlke, J, Lowenhaupt, K, Heinemann, U, Rich, A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins.
Nat.Struct.Biol., 8, 2001
|
|
1NRJ
 
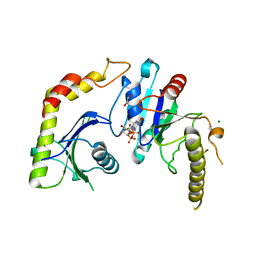 | |
1QBJ
 
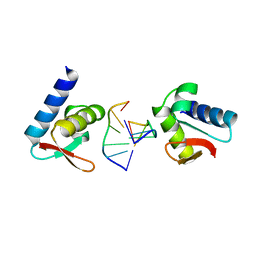 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
4KF7
 
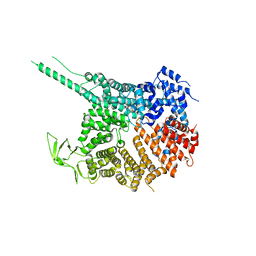 | |
4KF8
 
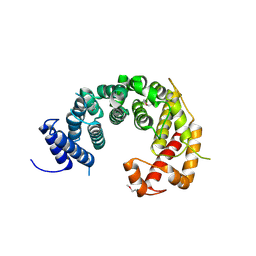 | |
4TVS
 
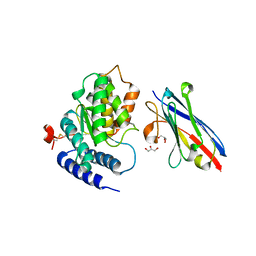 | | LAP1(aa356-583), H.sapiens, bound to VHH-BS1 | | Descriptor: | GLYCEROL, SULFATE ION, Torsin-1A-interacting protein 1, ... | | Authors: | Sosa, B.A, Schwartz, T.U. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Elife, 3, 2014
|
|
1XKS
 
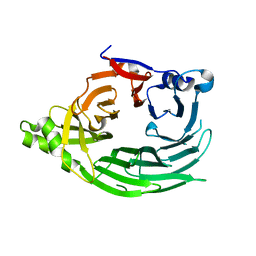 | |
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
2QX5
 
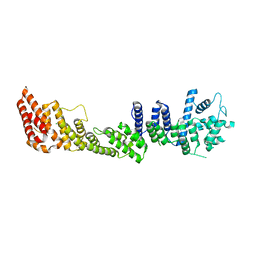 | | Structure of nucleoporin Nic96 | | Descriptor: | CHLORIDE ION, Nucleoporin NIC96 | | Authors: | Jeudy, S, Schwartz, T.U. | | Deposit date: | 2007-08-10 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture
J.Biol.Chem., 282, 2007
|
|
1BBA
 
 | |
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
5T0N
 
 | |
6OIF
 
 | | Cryo-EM structure of human TorsinA filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Torsin-1A | | Authors: | Zheng, W, Demircioglu, F.E, Schwartz, T.U, Egelman, E.H. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn.
Nat Commun, 10, 2019
|
|
5DJ4
 
 | |
2GED
 
 | |
5UK4
 
 | | VESICULAR STOMATITS VIRUS N PROTEIN IN COMPLEX WITH INHIBITORY NANOBODY 1307 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Nucleoprotein, RNA (45-MER) | | Authors: | Hanke, L, Knockenhauer, K.E, Ploegh, H.L, Schwartz, T.U. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication.
EMBO Rep., 18, 2017
|
|
5J1S
 
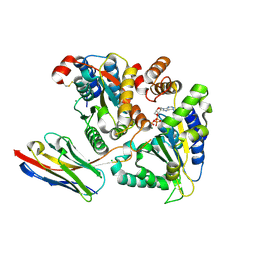 | | TorsinA-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5JJ7
 
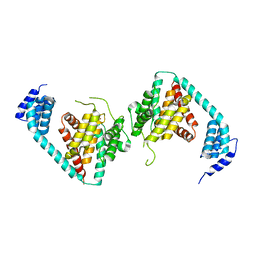 | | Fic-1 (aa134 - 508 E274G) from C. elegans | | Descriptor: | Adenosine monophosphate-protein transferase FICD homolog, SULFATE ION | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.749 Å) | | Cite: | Fic-1 (aa134 - 508 E274G) from C. elegans
To be published
|
|
5J1T
 
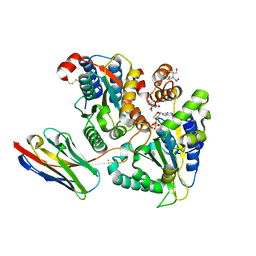 | | TorsinAdeltaE-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5JJ6
 
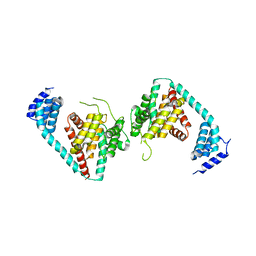 | |
3I4R
 
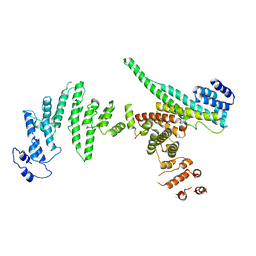 | | Nup107(aa658-925)/Nup133(aa517-1156) complex, H.sapiens | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-02 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
3CQC
 
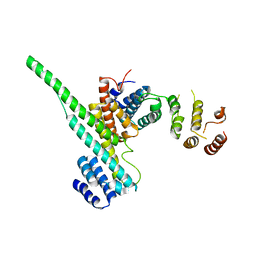 | | Nucleoporin Nup107/Nup133 interaction complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Jeudy, S, Boehmer, T, Berke, I, Schwartz, T.U. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Mol.Cell, 30, 2008
|
|
3CQG
 
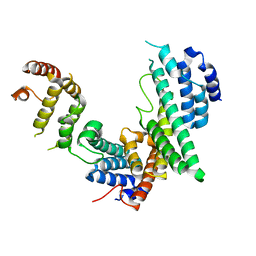 | | Nucleoporin Nup107/Nup133 interaction complex, delta finger mutant | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Jeudy, S, Boehmer, T, Berke, I, Schwartz, T.U. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Mol.Cell, 30, 2008
|
|
7TJC
 
 | | VHH Chl-B2 in complex with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, GLYCEROL, VHH-Chl-B2 | | Authors: | Nordeen, S.A, Schwartz, T.U. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and specificity of an anti-chloramphenicol single domain antibody for detection of amphenicol residues.
Protein Sci., 31, 2022
|
|
