5NK3
 
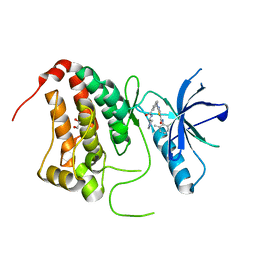 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1l | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{S})-pyrrolidin-3-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKC
 
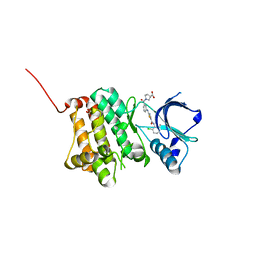 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2h | | Descriptor: | (3~{S})-1-[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKE
 
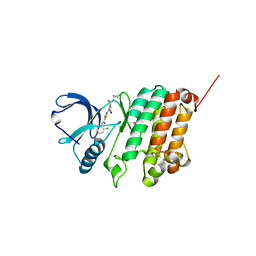 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3a | | Descriptor: | 2-[[3-bromanyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5N5C
 
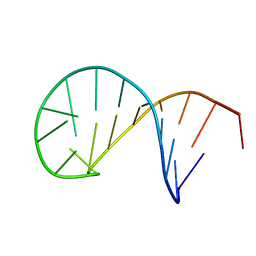 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
2JQ7
 
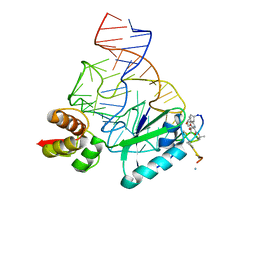 | | Model for thiostrepton binding to the ribosomal L11-RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RIBOSOMAL RNA, THIOSTREPTON | | Authors: | Jonker, H.R.A, Ilin, S, Grimm, S.K, Woehnert, J, Schwalbe, H. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy
Nucleic Acids Res., 35, 2007
|
|
2KOC
 
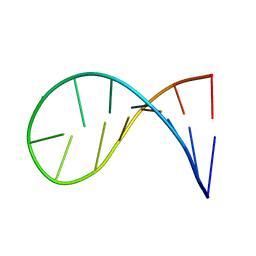 | | NMR solution structure of a 14-mer hairpin RNA with cUUCGg tetraloop | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nozinovic, S, Fuertig, B, Jonker, H.R.A, Richter, C, Schwalbe, H. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA
Nucleic Acids Res., 2009
|
|
2KER
 
 | | alpha-amylase inhibitor Parvulustat (Z-2685) from Streptomyces parvulus | | Descriptor: | Alpha-amylase inhibitor Z-2685 | | Authors: | Rehm, S, Han, S, Hassani, I, Sokocevic, A, Jonker, H.R.A, Engels, J.W, Schwalbe, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of parvulustat (Z-2685) from Streptomyces parvulus FH-1641: comparison with tendamistat from Streptomyces tendae 4158
Chembiochem, 10, 2009
|
|
2K5B
 
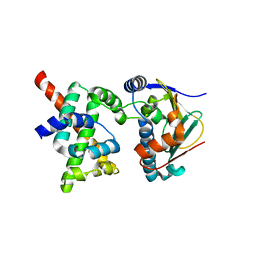 | | Human CDC37-HSP90 docking model based on NMR | | Descriptor: | Heat shock protein HSP 90-alpha, Hsp90 co-chaperone Cdc37 | | Authors: | Sreeramulu, S, Jonker, H.R.A, Lancaster, C.R, Richter, C, Langer, T, Schwalbe, H. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The human Cdc37.Hsp90 complex studied by heteronuclear NMR spectroscopy
J.Biol.Chem., 284, 2009
|
|
2KMJ
 
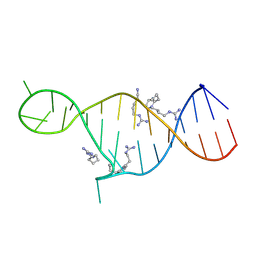 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
2L2Z
 
 | | Thiostrepton, reduced at N-CA bond of residue 14 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2W
 
 | | Thiostrepton | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L6X
 
 | | Solution NMR Structure of Proteorhodopsin. | | Descriptor: | Green-light absorbing proteorhodopsin, RETINAL | | Authors: | Reckel, S, Gottstein, D, Stehle, J, Loehr, F, Takeda, M, Silvers, R, Kainosho, M, Glaubitz, C, Bernhard, F, Schwalbe, H, Guntert, P, Doetsch, V, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-09 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of proteorhodopsin.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2LHP
 
 | |
2L2Y
 
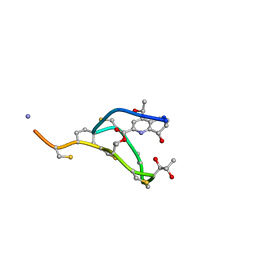 | | Thiostrepton, epimer form of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2X
 
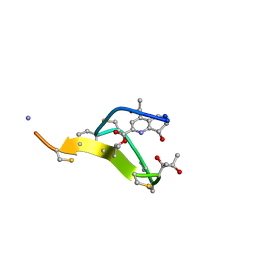 | | Thiostrepton, oxidized at CA-CB bond of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2LUB
 
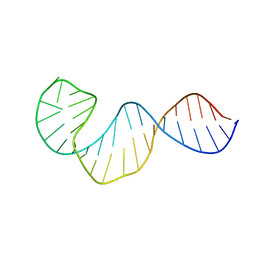 | |
2LUO
 
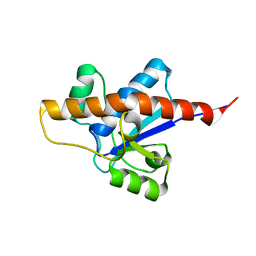 | | NMR solution structure of apo-MptpA | | Descriptor: | LOW MOLECULAR WEIGHT PROTEIN-TYROSINE-PHOSPHATASE A | | Authors: | Stehle, T, Sreeramulu, S, Loehr, F, Richter, C, Saxena, K, Jonker, H.R.A, Schwalbe, H. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Apo-structure of the Low Molecular Weight Protein-tyrosine Phosphatase A (MptpA) from Mycobacterium tuberculosis Allows for Better Target-specific Drug Development.
J.Biol.Chem., 287, 2012
|
|
2MW4
 
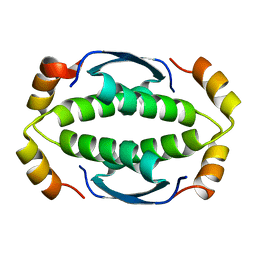 | | Tetramerization domain of the Ciona intestinalis p53/p73-b transcription factor protein | | Descriptor: | Transcription factor protein | | Authors: | Heering, J.P, Jonker, H.R.A, Loehr, F, Schwalbe, H, Doetsch, V. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain.
Protein Sci., 25, 2016
|
|
2N9Q
 
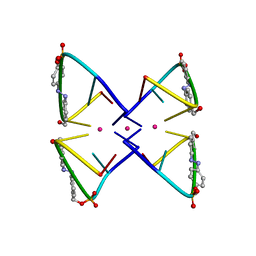 | | Photoswitchable G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*(AZW)P*GP*G)-3'), POTASSIUM ION | | Authors: | Thevarpadam, J, Bessi, I, Binas, O, Goncalves, D.P.N, Slavov, C, Jonker, H.R.A, Richter, C, Wachtveitl, J, Schwalbe, H, Heckel, A. | | Deposit date: | 2015-12-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photoresponsive Formation of an Intermolecular Minimal G-Quadruplex Motif.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5X1X
 
 | |
