4A69
 
 | | Structure of HDAC3 bound to corepressor and inositol tetraphosphate | | Descriptor: | ACETATE ION, D-MYO INOSITOL 1,4,5,6 TETRAKISPHOSPHATE, GLYCEROL, ... | | Authors: | Watson, P.J, Fairall, L, Santos, G.M, Schwabe, J.W.R. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Hdac3 Bound to Co-Repressor and Inositol Tetraphosphate.
Nature, 481, 2012
|
|
2XTC
 
 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XTD
 
 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4CP3
 
 | | The structure of BCL6 BTB (POZ) domain in complex with the ansamycin antibiotic rifabutin. | | Descriptor: | B-CELL LYMPHOMA 6 PROTEIN, RIFABUTIN | | Authors: | Evans, S.E, Fairall, L, Goult, B.T, Jamieson, A.G, Ferrigno, P.K, Ford, R, Wagner, S.D, Schwabe, J.W.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Ansamycin Antibiotic, Rifamycin Sv, Inhibits Bcl6 Transcriptional Repression and Forms a Complex with the Bcl6-Btb/Poz Domain.
Plos One, 9, 2014
|
|
7AZN
 
 | | Structure of mouse AsterC (GramD1c) with a new cholesterol-derived compound | | Descriptor: | 20alpha-hydroxy-20-(5-methylhexyl)cholesterol, ETHANOL, GLYCEROL, ... | | Authors: | Romartinez-Alonso, B, Sirvydis, K, Kim, Y, Xiao, X, Jung, M, Tontonoz, P, Schwabe, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Selective Aster inhibitors distinguish vesicular and nonvesicular sterol transport mechanisms.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2YHN
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MYLIP, ZINC ION | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
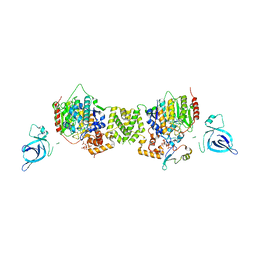 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
2XTE
 
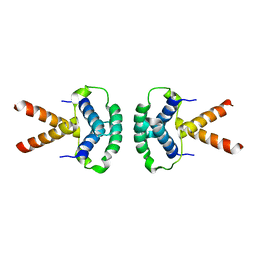 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2VV3
 
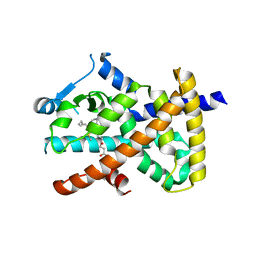 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VSR
 
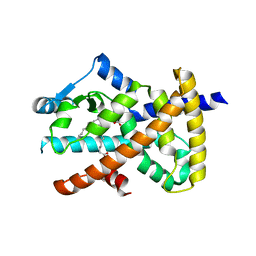 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV4
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxoOTE | | Descriptor: | (8E,10S,12Z)-10-hydroxy-6-oxooctadeca-8,12-dienoic acid, (8R,9Z,12Z)-8-hydroxy-6-oxooctadeca-9,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
