1JPH
 
 | | Ile260Thr mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPI
 
 | | Phe232Leu mutant of human UROD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JR2
 
 | | Structure of Uroporphyrinogen III Synthase | | Descriptor: | UROPORPHYRINOGEN-III SYNTHASE | | Authors: | Mathews, M.A, Schubert, H.L, Whitby, F.G, Alexander, K.J, Schadick, K, Bergonia, H.A, Phillips, J.D, Hill, C.P. | | Deposit date: | 2001-08-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of human uroporphyrinogen III synthase.
EMBO J., 20, 2001
|
|
1JPK
 
 | | Gly156Asp mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
3OYX
 
 | | Haloferax volcanii Malate Synthase Magnesium/Glyoxylate Complex | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3OYZ
 
 | | Haloferax volcanii Malate Synthase Pyruvate/Acetyl-CoA Ternary Complex | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3PUG
 
 | | Haloferax volcanii Malate Synthase Native at 3mM Glyoxylate | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C.D, Neighbor, A.M, Thomas, G.C, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-12-04 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
7MQO
 
 | | The insulin receptor ectodomain in complex with a venom hybrid insulin analog - "head" region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, ... | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQS
 
 | | The insulin receptor ectodomain in complex with three venom hybrid insulin molecules - asymmetric conformation | | Descriptor: | Insulin A chain, Insulin B chain, Isoform Short of Insulin receptor | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQR
 
 | | The insulin receptor ectodomain in complex with four venom hybrid insulins - symmetric conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
1S1Q
 
 | | TSG101(UEV) domain in complex with Ubiquitin | | Descriptor: | ACETIC ACID, COPPER (II) ION, SULFATE ION, ... | | Authors: | Sundquist, W.I, Schubert, H.L, Kelly, B.N, Hill, G.C, Holton, J.M, Hill, C.P. | | Deposit date: | 2004-01-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin recognition by the human TSG101 protein
Mol.Cell, 13, 2004
|
|
1S4D
 
 | | Crystal Structure Analysis of the S-adenosyl-L-methionine dependent uroporphyrinogen-III C-methyltransferase SUMT | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Vevodova, J, Graham, R.M, Raux, E, Schubert, H.L, Roper, D.I, Brindley, A.A, Scott, A.I, Roessner, C.A, Stamford, N.P.J, Stroupe, M.E, Getzoff, E.D, Warren, M.J, Wilson, K.S. | | Deposit date: | 2004-01-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure/Function Studies on a S-Adenosyl-l-methionine-dependent Uroporphyrinogen III C Methyltransferase (SUMT), a Key Regulatory Enzyme of Tetrapyrrole Biosynthesis
J.Mol.Biol., 344, 2004
|
|
3FRV
 
 | |
1YPT
 
 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
1XXP
 
 | | Yersinia YopH (residues 163-468) C403S binds phosphotyrosyl peptide at two sites | | Descriptor: | Hexapeptide ASP-ALA-ASP-GLU-PTR-CLE, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
1XXV
 
 | | Yersinia YopH (residues 163-468) binds phosphonodifluoromethyl-Phe containing hexapeptide at two sites | | Descriptor: | Epidermal growth factor receptor derived peptide, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
1Z3Y
 
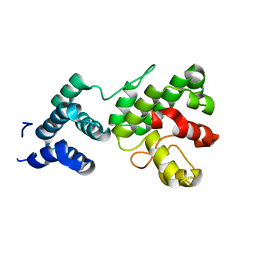 | | Structure of Gun4-1 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z3X
 
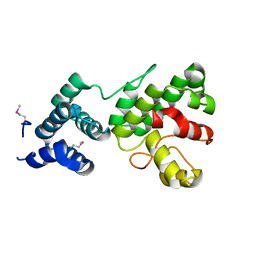 | | Structure of Gun4 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1ZY7
 
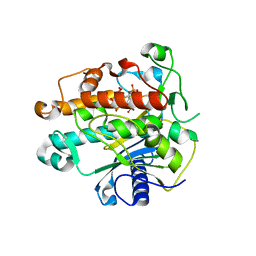 | | Crystal structure of the catalytic domain of an adenosine deaminase that acts on RNA (hADAR2) bound to inositol hexakisphosphate (IHP) | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RNA-specific adenosine deaminase B1, isoform DRADA2a, ... | | Authors: | Macbeth, M.R, Schubert, H.L, Vandemark, A.P, Lingam, A.T, Hill, C.P, Bass, B.L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing.
Science, 309, 2005
|
|
