4IE5
 
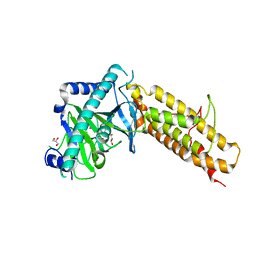 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-[(3-hydroxypyridin-2-yl)carbonyl]glycine (MD6) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-[(3-hydroxypyridin-2-yl)carbonyl]glycine, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4IE7
 
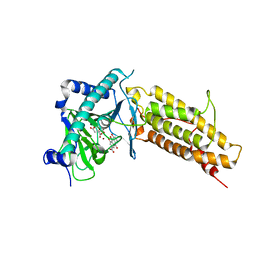 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with citrate and rhein (RHN) | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, CITRATE ANION, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6004 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4IE6
 
 | |
4IDZ
 
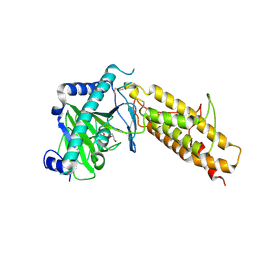 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-oxalylglycine (NOG) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4IE4
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5048 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
6Y0O
 
 | | isopenicillin N synthase in complex with ACV and Fe under anaerobic environment using FT-SSX methods | | Descriptor: | Anaerobic Fixed Target Structure of Isopenicillin N synthase in complex with Fe and ACV, FE (III) ION, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
2QC9
 
 | |
6YPV
 
 | | Alpha-ketoglutarate-dependent dioxygenase AlkB in complex with Fe and AKG after oxygen exposure using FT-SSX methods | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
2JE5
 
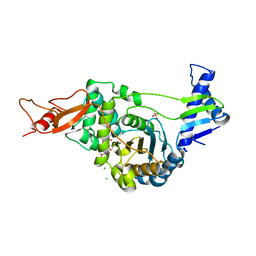 | | STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING PROTEIN INHIBITION BY LACTIVICINS | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
6Y0Q
 
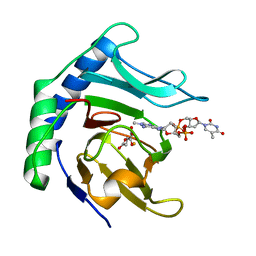 | | Alpha-ketoglutarate-dependent dioxygenase AlkB in complex with Fe, AKG and methylated DNA under anaerobic environment using FT-SSX methods | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
1H2K
 
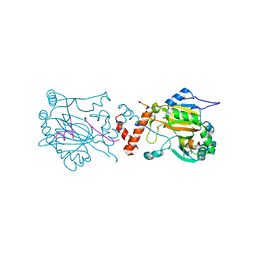 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2M
 
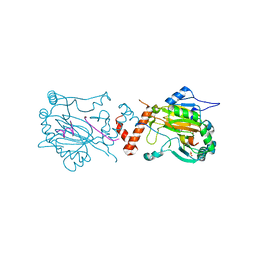 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, N-OXALYLGLYCINE, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
6Y6J
 
 | | VIM-2 in Complex with Biapenem Imine and Enamine Hydrolysis Products | | Descriptor: | Biapenem Enamine hydrolysis product, Biapenem Imine hydrolysis product, CHLORIDE ION, ... | | Authors: | Lucic, A, Schofield, C.J, Brem, J, McDonough, M.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of metallo-beta-lactamase VIM-2 in complex with the imine and enamine form of hydrolysed Biapenem
To Be Published
|
|
1H2L
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FACTOR INHIBITING HIF1, FE (II) ION, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
2A1X
 
 | | Human phytanoyl-coa 2-hydroxylase in complex with iron and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Phytanoyl-CoA dioxygenase | | Authors: | Kavanagh, K.L, McDonough, M.A, Searles, T, Butler, D, Bunkoczi, G, von Delft, F, Edwards, A, Arrowsmith, C, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Phytanoyl-CoA 2-Hydroxylase Identifies Molecular Mechanisms of Refsum Disease
J.Biol.Chem., 280, 2005
|
|
2OS2
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys36 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OT7
 
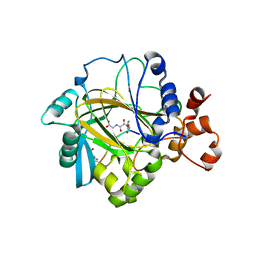 | | Crystal structure of JMJD2A complexed with histone H3 peptide monomethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OQ6
 
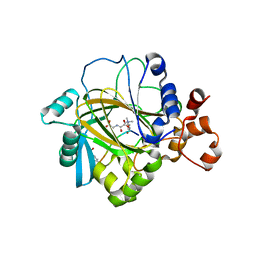 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OQ7
 
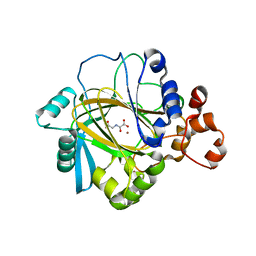 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
1H2N
 
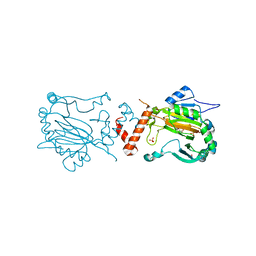 | | Factor Inhibiting HIF-1 alpha | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
8C8B
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 48). | | Descriptor: | 4-[[(2~{S})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-~{N}-prop-2-enyl-quinazoline-2-carboxamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21).
To Be Published
|
|
8C8D
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 44). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(furan-2-ylmethylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 44).
To Be Published
|
|
8C8S
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(prop-2-enylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21).
To Be Published
|
|
2G1M
 
 | |
2WOL
 
 | | Clavulanic acid biosynthesis oligopeptide binding protein 2 | | Descriptor: | CLAVULANIC ACID BIOSYNTHESIS OLIGOPEPTIDE BINDING PROTEIN 2, GLYCEROL | | Authors: | MacKenzie, A.K, Valegard, K, Iqbal, A, Caines, M.E.C, Kershaw, N.J, Jensen, S.E, Schofield, C.J, Andersson, I. | | Deposit date: | 2009-07-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of an Oligopeptide-Binding Protein from the Biosynthetic Pathway of the Beta-Lactamase Inhibitor Clavulanic Acid.
J.Mol.Biol., 396, 2010
|
|
