8VI2
 
 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
7UYD
 
 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYA
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
1HMT
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, STEARIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HMR
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | 9-OCTADECENOIC ACID, MUSCLE FATTY ACID BINDING PROTEIN | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HMS
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
2HMB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN MUSCLE FATTY ACID-BINDING PROTEIN | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Zanotti, G, Scapin, G, Spadon, P, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1992-09-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of recombinant human muscle fatty acid-binding protein.
J.Biol.Chem., 267, 1992
|
|
1B9I
 
 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE) | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
1B9H
 
 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
7MD4
 
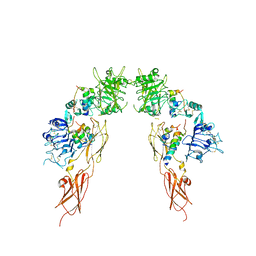 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
7MD5
 
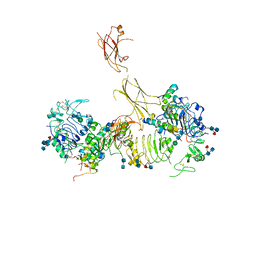 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
1HMP
 
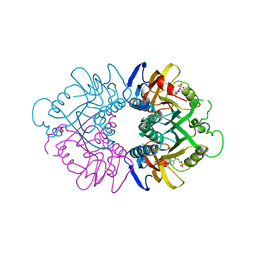 | | THE CRYSTAL STRUCTURE OF HUMAN HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH BOUND GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE GUANINE PHOSPHORIBOSYL-TRANSFERASE | | Authors: | Eads, J.C, Scapin, G, Xu, Y, Grubmeyer, C, Sacchettini, J.C. | | Deposit date: | 1994-06-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP.
Cell(Cambridge,Mass.), 78, 1994
|
|
7LEX
 
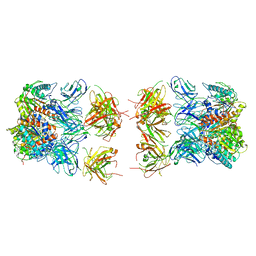 | |
1C3V
 
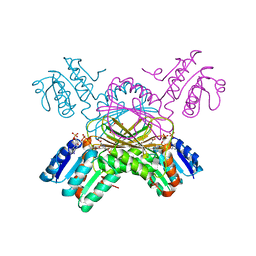 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
1LQF
 
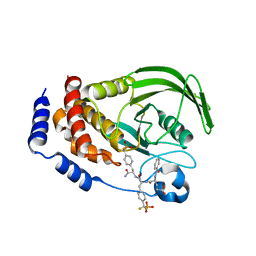 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | Descriptor: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | Deposit date: | 2002-05-10 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
7LEY
 
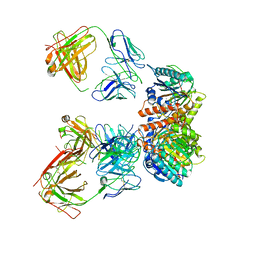 | | Trimeric human Arginase 1 in complex with mAb5 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF1
 
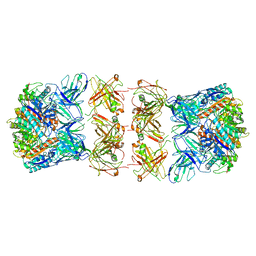 | | Trimeric human Arginase 1 in complex with mAb3 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF2
 
 | | Trimeric human Arginase 1 in complex with mAb4 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEZ
 
 | | Trimeric human Arginase 1 in complex with mAb1 - 2 hArg:2 mAb1 complex | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb1 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF0
 
 | |
1OVE
 
 | | The structure of p38 alpha in complex with a dihydroquinolinone | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERIDIN-4-YL-3,4-DIHYDROQUINOLIN-2(1H)-ONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
