1K4Q
 
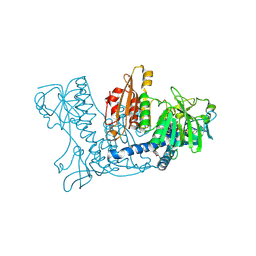 | | Human Glutathione Reductase Inactivated by Peroxynitrite | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione Reductase | | Authors: | Savvides, S.N, Scheiwein, M, Boehme, C.C, Arteel, G.E, Karplus, P.A, Becker, K, Schirmer, R.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite.
J.Biol.Chem., 277, 2002
|
|
1ETE
 
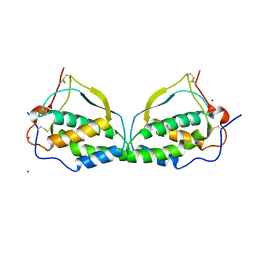 | |
1NLY
 
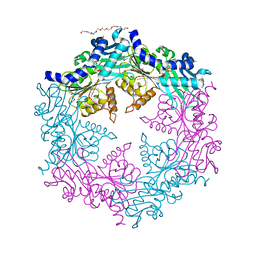 | | Crystal structure of the traffic ATPase of the Helicobacter pylori type IV secretion system in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-06 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
7OX6
 
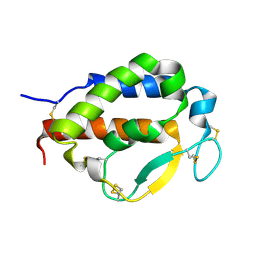 | | Solution structure of human interleukin-9 | | Descriptor: | Interleukin-9 | | Authors: | Savvides, S.N, Tripsianes, K, De Vos, T, Papageorgiou, A, Evangelidis, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-11 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
1SRU
 
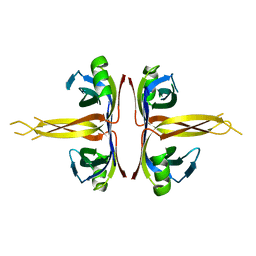 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1OPX
 
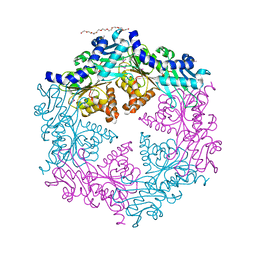 | | Crystal structure of the traffic ATPase (HP0525) of the Helicobacter pylori type IV secretion system bound by sulfate | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1NLZ
 
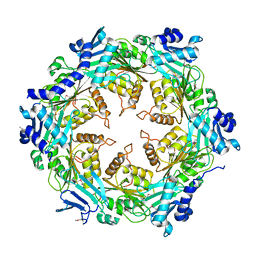 | | Crystal structure of unliganded traffic ATPase of the type IV secretion system of helicobacter pylori | | Descriptor: | virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
2I9V
 
 | |
1XAN
 
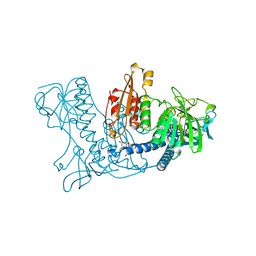 | |
2GQ8
 
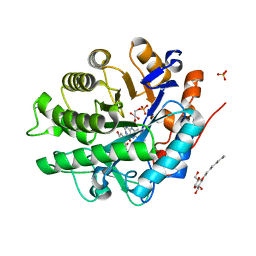 | | Structure of SYE1, an OYE homologue from S. ondeidensis, in complex with p-hydroxyacetophenone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, P-HYDROXYACETOPHENONE, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
2GOU
 
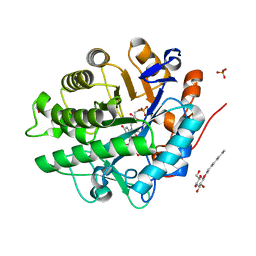 | | Structure of wild type, oxidized SYE1, an OYE homologue from S. oneidensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
2GQA
 
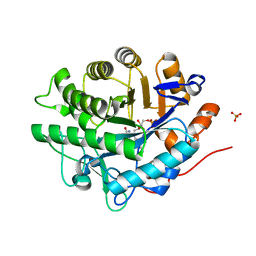 | | Structure of NADH-reduced SYE1, an OYE homologue from S. oneidensis | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, oxidoreductase, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
2GQ9
 
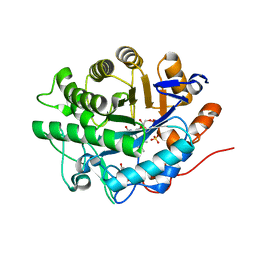 | | Structure of SYE1, an OYE homologue from S. oneidensis, in complex with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, SULFATE ION, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
6E4L
 
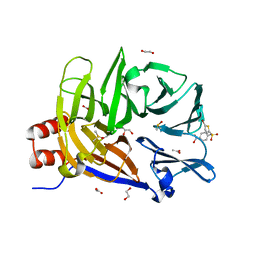 | | The structure of the N-terminal domain of human clathrin heavy chain 1 (nTD) in complex with ES9 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(4-nitrophenyl)thiophene-2-sulfonamide, ACETATE ION, ... | | Authors: | Dejonghe, W, Sharma, I, Denoo, B, Munck, S.D, Bulut, H, Mylle, E, Vasileva, M, Lu, Q, Savatin, D.V, Mishev, K, Nerinckx, W, Staes, A, Drozdzecki, A, Audenaert, D, Madder, A, Friml, J, Damme, D.V, Gevaert, K, Haucke, V, Savvides, S, Winne, J, Russinova, E. | | Deposit date: | 2018-07-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of endocytosis through chemical inhibition of clathrin heavy chain function.
Nat.Chem.Biol., 15, 2019
|
|
8C7L
 
 | |
8C7E
 
 | |
3UCE
 
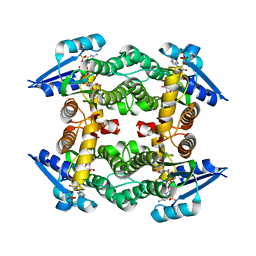 | | Crystal structure of a small-chain dehydrogenase in complex with NADPH | | Descriptor: | Dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Buysschaert, G, Verstraete, K, Savvides, S, Vergauwen, B. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference.
Febs J., 280, 2013
|
|
3LN7
 
 | |
7NX2
 
 | | Unbound antigen-binding fragment (FAb) 324 | | Descriptor: | FAb 324 Heavy Chain, FAb 324 Light Chain, SULFATE ION | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
7NX4
 
 | |
7NWZ
 
 | | ALK:ALKAL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
7NX3
 
 | | Crystal structure of ALK in complex with Fab324 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor, Fab324 HeavyChain, ... | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
7NX0
 
 | | LTK:ALKAL1 complex stabilized by a Nanobody | | Descriptor: | ALK and LTK ligand 1, Leukocyte tyrosine kinase receptor, Nb3.16, ... | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
7NX1
 
 | | TG domain of LTK | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leukocyte tyrosine kinase receptor, TERBIUM(III) ION | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
8C6Z
 
 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
