4P77
 
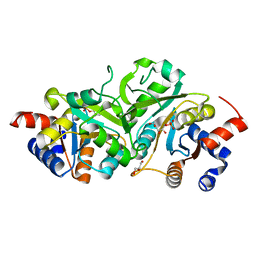 | | Structure of ribB complexed with substrate Ru5P | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, RIBULOSE-5-PHOSPHATE | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6D
 
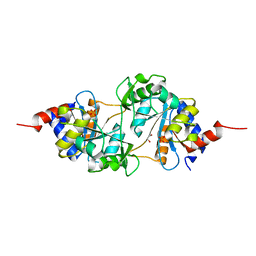 | | Structure of ribB complexed with PO4 ion | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8J
 
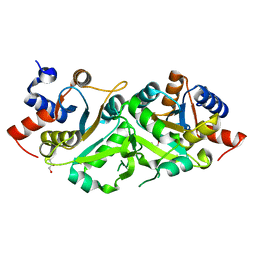 | | Structure of ribB | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
2Q1D
 
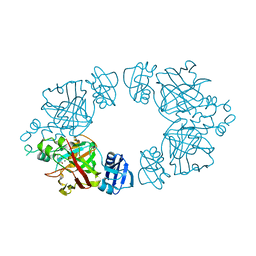 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with magnesium and 2,5-dioxopentanoate | | Descriptor: | 2,5-DIOXOPENTANOIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, MAGNESIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q1C
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with calcium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, CALCIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q19
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase apo form | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q18
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase, PHOSPHATE ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q1A
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with magnesium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, MAGNESIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2X1S
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1R
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-(PROPYLSULFONYL)PROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2MCN
 
 | | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Salmon, L, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications.
Plos One, 8, 2013
|
|
1HM5
 
 | |
