6QIV
 
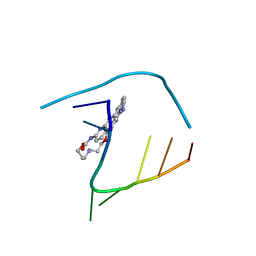 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL4 compound | | Descriptor: | CMBL4, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIQ
 
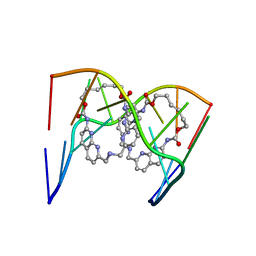 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIT
 
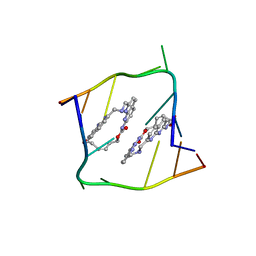 | | Crystal structure of CAG repeats with synthetic CMBL3b compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIR
 
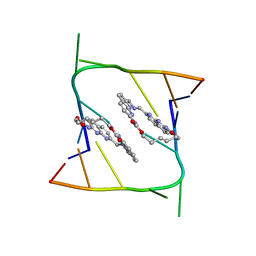 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
1LNL
 
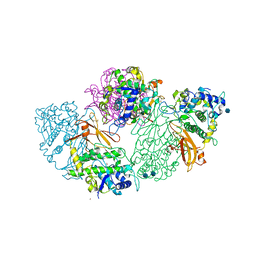 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
1M2T
 
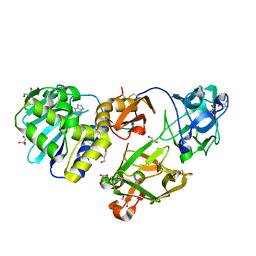 | | Mistletoe Lectin I from Viscum album in Complex with Adenine Monophosphate. Crystal Structure at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Krauspenhaar, R, Rypniewski, W, Kalkura, N, Moore, K, DeLucas, L, Stoeva, S, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallisation under microgravity of mistletoe lectin I from Viscum album with adenine monophosphate and the crystal structure at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3GM7
 
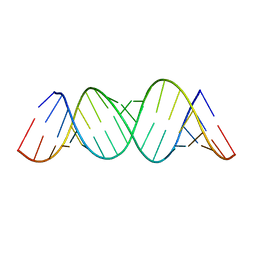 | |
3GLP
 
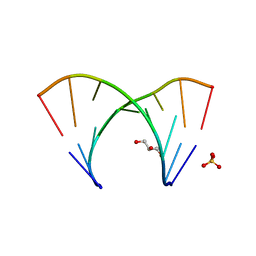 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
5EME
 
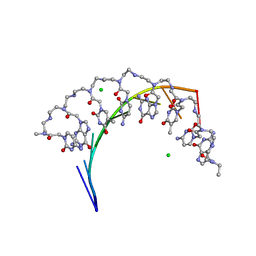 | | Complex of RNA r(GCAGCAGC) with antisense PNA p(CTGCTGC) | | Descriptor: | Antisense PNA strand, CHLORIDE ION, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
5EMG
 
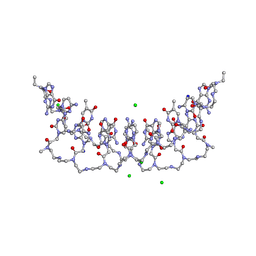 | | Crystal structures of PNA p(GCTGCTGC)2 duplex containing T-T mismatches | | Descriptor: | CHLORIDE ION, GPN-CPN-TPN-GPN-CPN-TPN-GPN-CPN, SODIUM ION | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
5EMF
 
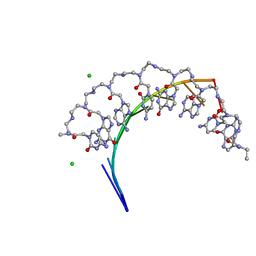 | | Crystal structure of RNA r(GCUGCUGC) with antisense PNA p(GCAGCAGC) | | Descriptor: | CHLORIDE ION, RNA (5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3'), antisense PNA p(GCAGCAGC) | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
6HNU
 
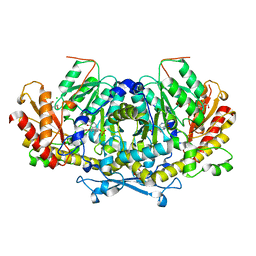 | | Crystal structure of the aminotransferase Aro8 from C. Albicans with ligands | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
6HNB
 
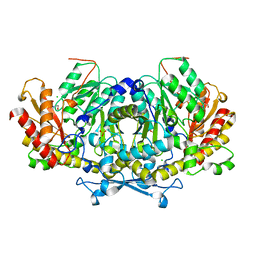 | | Crystal structure of aminotransferase Aro8 from Candida albicans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, CHLORIDE ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
6HND
 
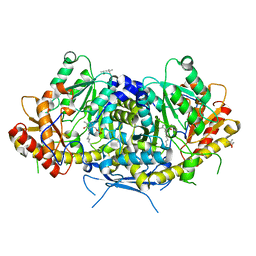 | | Crystal structure of the aromatic aminotransferase Aro9 from C. Albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Aromatic-amino-acid:2-oxoglutarate transaminase, POTASSIUM ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
6HNV
 
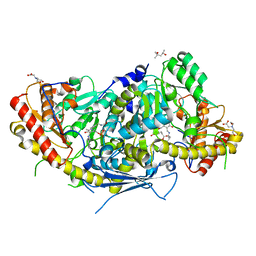 | | Crystal structure of aminotransferase Aro9 from C. Albicans with ligands | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINOHEXANEDIOIC ACID, 2-OXOADIPIC ACID, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
4EZ8
 
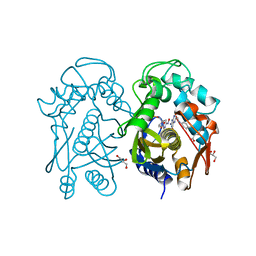 | | Crystal structure of mouse thymidylate sythase in ternary complex with N(4)-hydroxy-2'-deoxycytidine-5'-monophosphate and the cofactor product, dihydrofolate | | Descriptor: | 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), DIHYDROFOLIC ACID, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Wilk, P, Kierdaszuk, B, Banaszak, K, Gorecka, K, Rode, W. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structures of complexes of mouse thymidylate synthase
crystallized with N4-OH-dCMP alone or in the presence of
N5,10-methylenetetrahydrofolate
Pteridines, 2013
|
|
2POC
 
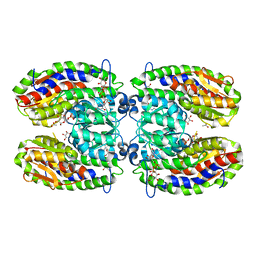 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
4MB4
 
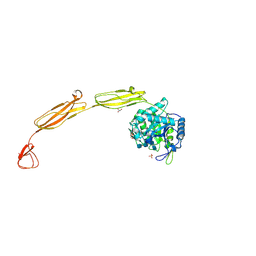 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
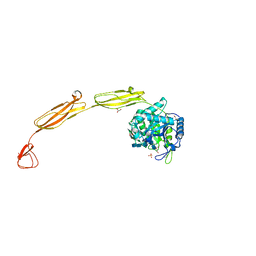 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
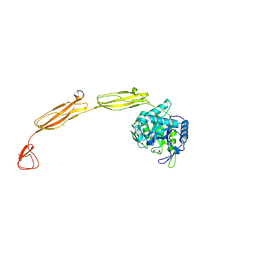 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IRR
 
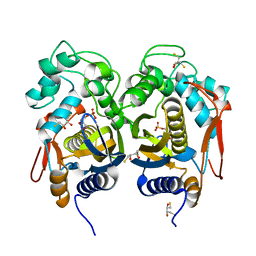 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
4IQB
 
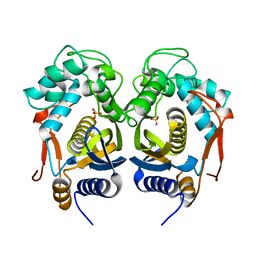 | | High Resolution Crystal Structure of C.elegans Thymidylate Synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
4ISW
 
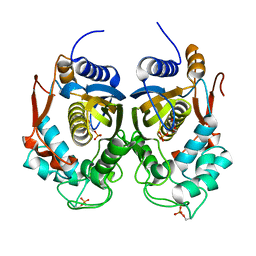 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
3O8L
 
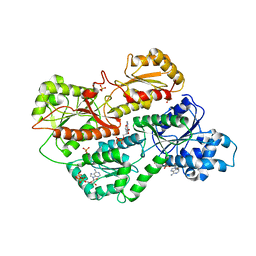 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3R1C
 
 | | Crystal structure of GCGGCGGC duplex | | Descriptor: | RNA (5'-R(*GP*CP*GP*GP*CP*GP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0531 Å) | | Cite: | Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome.
Nucleic Acids Res., 39, 2011
|
|
