6S9F
 
 | | Drosophila OTK, extracellular domains 3-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tyrosine-protein kinase-like otk | | Authors: | Rozbesky, D, Jones, E.Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.
Structure, 28, 2020
|
|
6QP9
 
 | | Drosophila Semaphorin 1a, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-1A | | Authors: | Rozbesky, D, Robinson, R.A, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6FKM
 
 | |
6FKK
 
 | | Drosophila Semaphorin 1b, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MIP07328p, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rozbesky, D, Harlos, K, Jones, E.Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6FKN
 
 | |
8RMJ
 
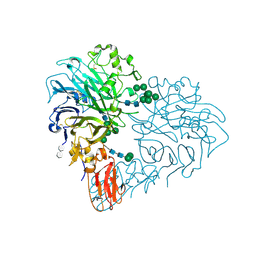 | | Drosophila Semaphorin 2b in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nourisanami, F, Sobol, M, Rozbesky, D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanisms of proteoglycan-mediated semaphorin signaling in axon guidance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3M9Z
 
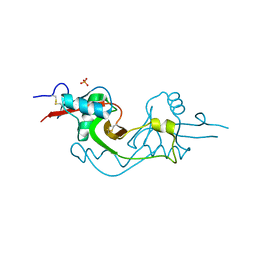 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
7BH8
 
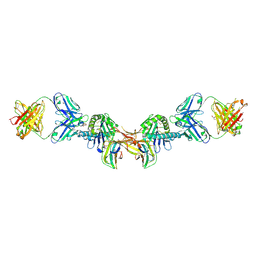 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
3T3A
 
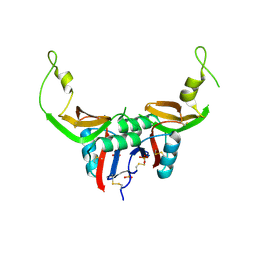 | | Crystal structure of H107R mutant of extracellular domain of mouse receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2M94
 
 | |
2MTI
 
 | | NMR structure of the lymphocyte receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A | | Authors: | Chmelik, J, Rozbesky, D, Pospisilova, E, Adamek, D, Novak, P. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lymphocyte receptor Nkrp1a reveals a distinct conformation of the long loop region as compared to in the crystal structure.
Proteins, 84, 2016
|
|
6QP7
 
 | | Drosophila Semaphorin 2a | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6QP8
 
 | | Drosophila Semaphorin 2b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6GGM
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Phe. | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Mtb44*P2-Phe peptide variant (ARG-PHE-PRO-ALA-LYS-ALA-PRO-LEU-LEU), ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH4
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Gln. | | Descriptor: | ARG-GLN-PRO-ALA-LYS-ALA-PRO-LEU-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GL1
 
 | | HLA-E*01:03 in complex with the HIV epitope, RL9HIV | | Descriptor: | ARG-MET-TYR-SER-PRO-THR-SER-ILE-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH1
 
 | | HLA-E*01:03 in complex with Mtb44 | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GHN
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P9-Phe. | | Descriptor: | ARG-LEU-PRO-ALA-LYS-ALA-PRO-LEU-PHE, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
