3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
8G0I
 
 | |
8SXU
 
 | |
8SXT
 
 | | Structure of LINE-1 ORF2p with template:primer hybrid | | Descriptor: | DNA primer, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | van Eeuwen, T, Taylor, M.S, Rout, M.P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
8SFV
 
 | | High affinity nanobodies to GFP | | Descriptor: | GLYCEROL, Green fluorescent protein, LaG19, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High affinity nanobodies to GFP
To Be Published
|
|
8SFS
 
 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SLC
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8TJ5
 
 | | Inner spoke ring of the yeast NPC | | Descriptor: | Nucleoporin 59, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
8SFZ
 
 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8TIE
 
 | | Double nuclear outer ring of Nup84-complexes from the yeast NPC | | Descriptor: | NUP133 isoform 1, NUP145 isoform 1, Nucleoporin NUP120, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-19 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
8SG3
 
 | |
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
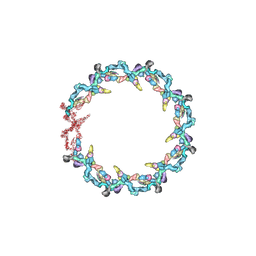 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
8T9L
 
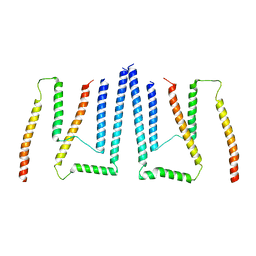 | | Pom34-Pom152 membrane attachment site yeast NPC | | Descriptor: | Nucleoporin POM152, Nucleoporin POM34 | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-06-24 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
2M4M
 
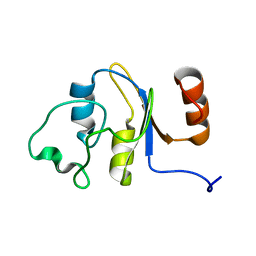 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
3NF5
 
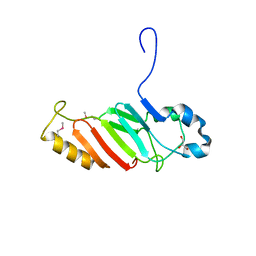 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
8SFX
 
 | | High Affinity nanobodies against GFP | | Descriptor: | D-MALATE, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
4OO6
 
 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
2MKG
 
 | |
2MKF
 
 | |
4JLQ
 
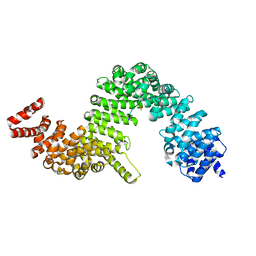 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
