1GQ7
 
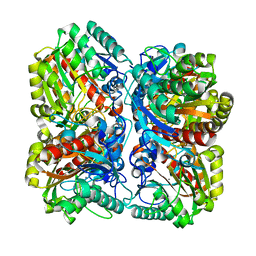 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oligomeric structure of proclavaminic acid amidino hydrolase: evolution of a hydrolytic enzyme in clavulanic acid biosynthesis.
Biochem. J., 366, 2002
|
|
1GQ6
 
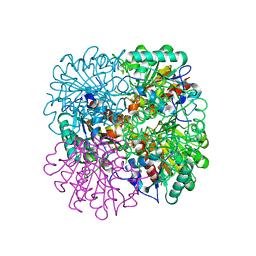 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric Structure of Proclavaminic Acid Amidino Hydrolase: Evolution of a Hydrolytic Enzyme in Clavulanic Acid Biosynthesis
Biochem.J., 366, 2002
|
|
2ARV
 
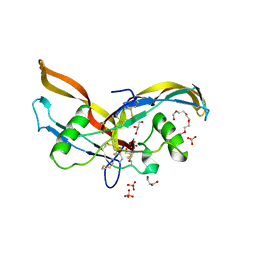 | | Structure of human Activin A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Inhibin beta A chain, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
8OX0
 
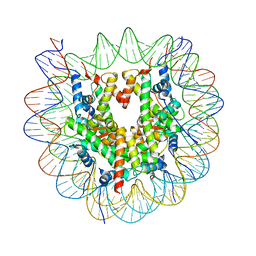 | | Structure of apo telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
8OX1
 
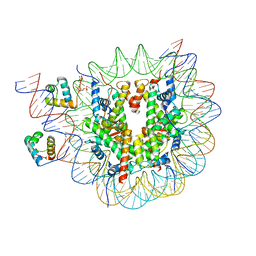 | | Structure of TRF1core in complex with telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
3W03
 
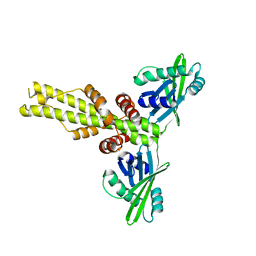 | | XLF-XRCC4 complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (8.492 Å) | | Cite: | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
6O7T
 
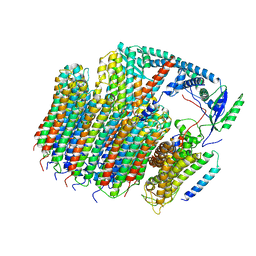 | | Saccharomyces cerevisiae V-ATPase Vph1-VO | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7V
 
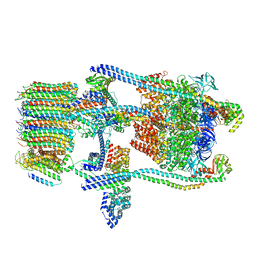 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7W
 
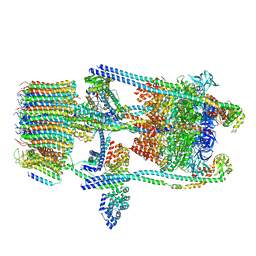 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 2 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7X
 
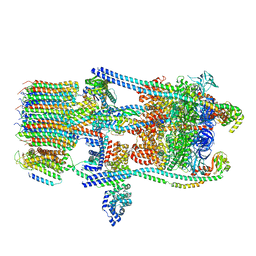 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7U
 
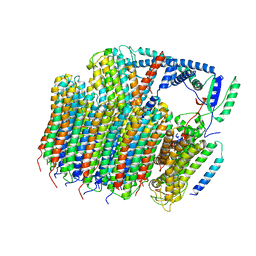 | | Saccharomyces cerevisiae V-ATPase Stv1-VO | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit a, Golgi isoform, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5EXG
 
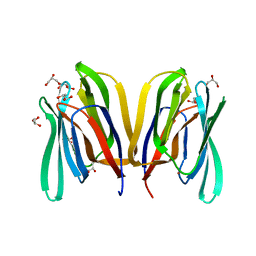 | |
7AIR
 
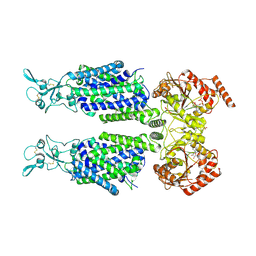 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Subclass 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Liko, I, Tehan, B.G, Almeida, F.G, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIQ
 
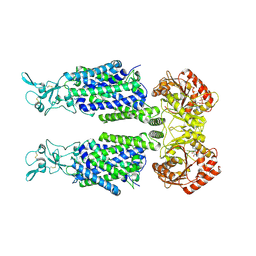 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Subclass 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 4, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Liko, I, Tehan, B.G, Almeida, F.G, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIP
 
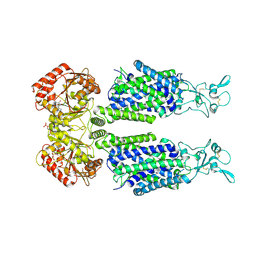 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Liko, I, Tehan, B.G, Almeida, F.G, Elkins, J, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
4NH2
 
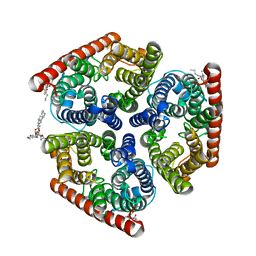 | | Crystal structure of AmtB from E. coli bound to phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Ammonia channel | | Authors: | Laganowsky, A, Reading, E, Allison, T.M, Robinson, C.V. | | Deposit date: | 2013-11-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membrane proteins bind lipids selectively to modulate their structure and function.
Nature, 510, 2014
|
|
5TJ5
 
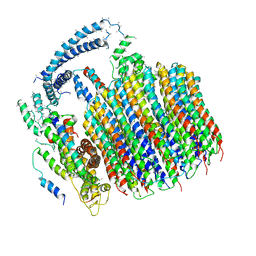 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
5ADX
 
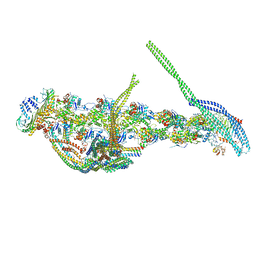 | | CryoEM structure of dynactin complex at 4.0 angstrom resolution | | Descriptor: | ACTIN RELATED PROTEIN 1, ACTIN RELATED PROTEIN 11, ACTIN, ... | | Authors: | Zhang, K, Urnavicius, L, Diamant, A.G, Motz, C, Schlage, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-08-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
5AFR
 
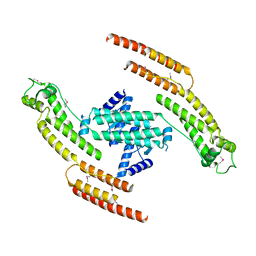 | | N-terminal fragment of dynein heavy chain | | Descriptor: | DYNEIN HEAVY CHAIN, CYTOPLASMIC | | Authors: | Urnavicius, L, Zhang, K, Diamant, A.G, Motz, C, Schlager, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
5AFU
 
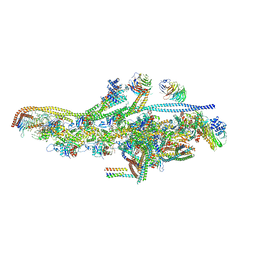 | | Cryo-EM structure of dynein tail-dynactin-BICD2N complex | | Descriptor: | ACTIN, CYTOPLASMIC 1, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Urnavicius, L, Zhang, K, Diamant, A.G, Motz, C, Schlager, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-01-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
4I0C
 
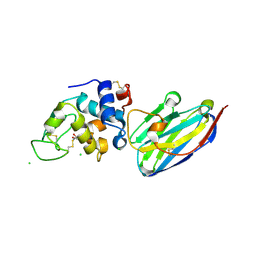 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
4M5T
 
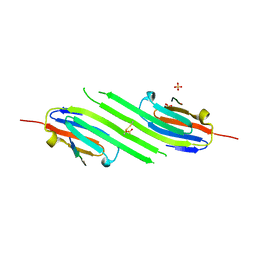 | | Disulfide trapped human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Cascio, D, Hochberg, G, Sawaya, M.R, Benesch, J.L.P, Robinson, C.V, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3D36
 
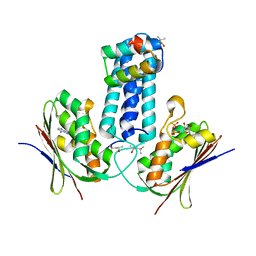 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
5FQ8
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-decanoyloxypropyl decanoate, BT_2262 (UNCHARACTERISED LIPOPROTEIN), ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ6
 
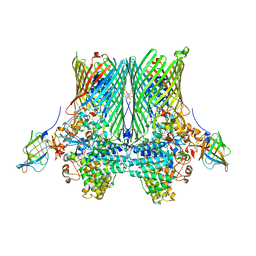 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | 3-decanoyloxypropyl decanoate, BT_2261, CALCIUM ION, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
