1Q6R
 
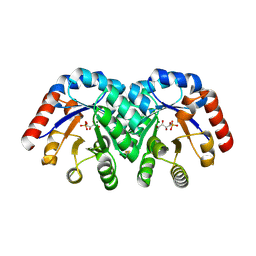 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-xylulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1QWG
 
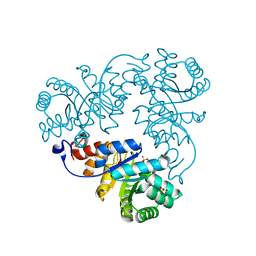 | | Crystal structure of Methanococcus jannaschii phosphosulfolactate synthase | | Descriptor: | (2R)-phospho-3-sulfolactate synthase, SULFATE ION | | Authors: | Wise, E.L, Graham, D.E, White, R.H, Rayment, I. | | Deposit date: | 2003-09-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural determination of phosphosulfolactate synthase from Methanococcus jannaschii at 1.7-A resolution: an enolase that is not an enolase
J.Biol.Chem., 278, 2003
|
|
1Q6L
 
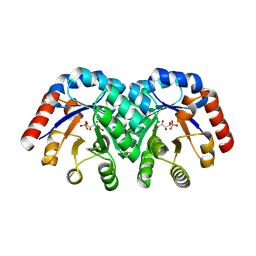 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1P90
 
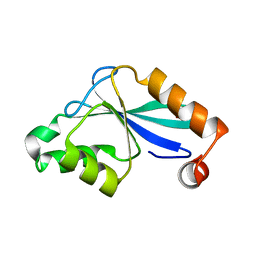 | | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 resolution | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein | | Authors: | Dyer, D.H, Rubio, L.M, Thoden, J.B, Holden, H.M, Ludden, P.W, Rayment, I. | | Deposit date: | 2003-05-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 A resolution
J.Biol.Chem., 278, 2003
|
|
1EC7
 
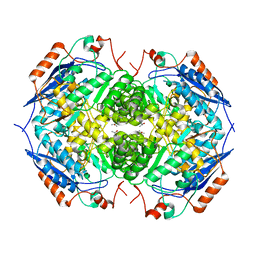 | | E. COLI GLUCARATE DEHYDRATASE NATIVE ENZYME | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1ECQ
 
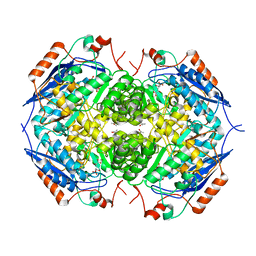 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO 4-DEOXYGLUCARATE | | Descriptor: | 4-DEOXYGLUCARATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EC8
 
 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO PRODUCT 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1BQG
 
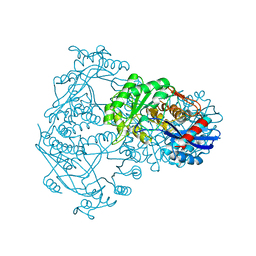 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
1EC9
 
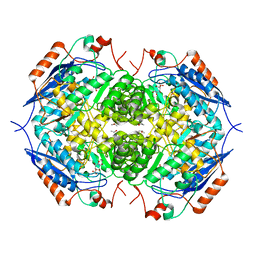 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1JDF
 
 | | Glucarate Dehydratase from E.coli N341D mutant | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JCT
 
 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1EBH
 
 | |
1TLK
 
 | |
1JDB
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Wesenberg, G, Raushel, F.M, Rayment, I. | | Deposit date: | 1997-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1TUW
 
 | | Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A Type-II Polyketide Cyclase | | Descriptor: | SULFATE ION, Tetracenomycin polyketide synthesis protein tcmI | | Authors: | Thompson, T.B, Katayama, K, Watanabe, K, Hutchinson, C.R, Rayment, I. | | Deposit date: | 2004-06-25 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase.
J.Biol.Chem., 279, 2004
|
|
1D0Z
 
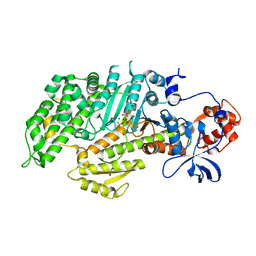 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH P-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, P-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1B
 
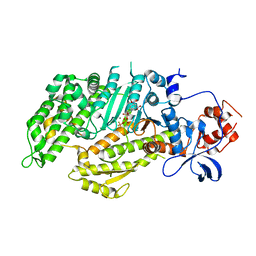 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, O,P-DINITROPHENYL AMINOPROPYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1C
 
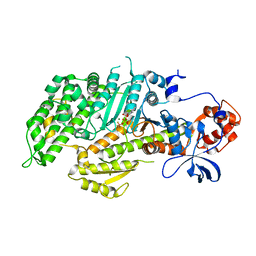 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH N-METHYL-O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, N-METHYL O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D1A
 
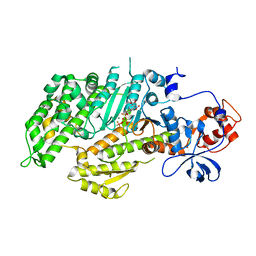 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O,P-DINITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, O,P-DINITROPHENYL AMINOETHYLDIPHOSPHATE-BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1MMA
 
 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
1MMG
 
 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
1D0X
 
 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH M-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | M-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE, MAGNESIUM ION, MYOSIN S1DC MOTOR DOMAIN | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1D0Y
 
 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM FLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN S1DC MOTOR DOMAIN, O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1EBG
 
 | |
1LC7
 
 | | Crystal Structure of L-Threonine-O-3-phosphate Decarboxylase from S. enterica complexed with a substrate | | Descriptor: | L-Threonine-O-3-Phosphate Decarboxylase, PHOSPHATE ION, PHOSPHOTHREONINE | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-04-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
Biochemistry, 41, 2002
|
|
