2FMX
 
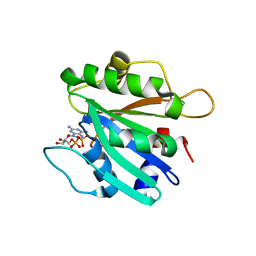 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
2FA9
 
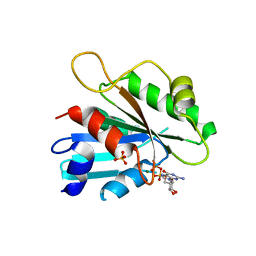 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
4N41
 
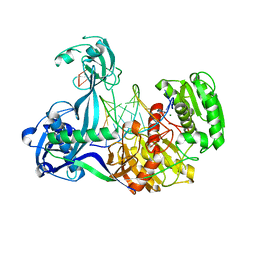 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N47
 
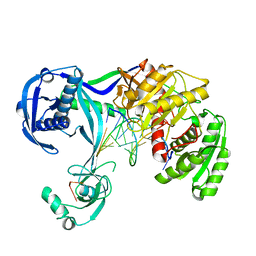 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCA
 
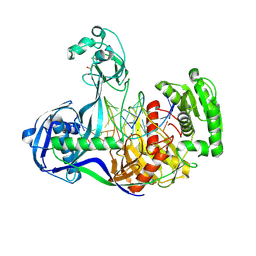 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | Descriptor: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCB
 
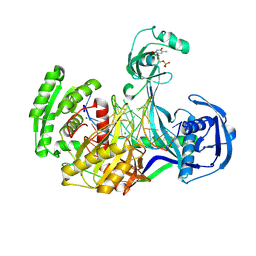 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 19-mer target DNA with Mg2+ | | Descriptor: | 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3', 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8H5D
 
 | |
8HSN
 
 | |
8HUI
 
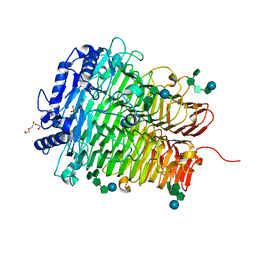 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 in complex with GF4, DFA I, and fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 in complex with GF4, DFA I, and fructose
To Be Published
|
|
5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
6KZS
 
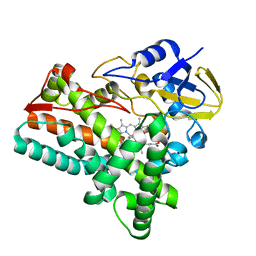 | | Crystal structure of cytochrome P450mel 107F1 in complex with heme and imidazole | | Descriptor: | Cytochrome P-450, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
6KZT
 
 | | Crystal structure of cytochrome P450mel 107F1 with biaryl coupling reactivity | | Descriptor: | Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
8HFJ
 
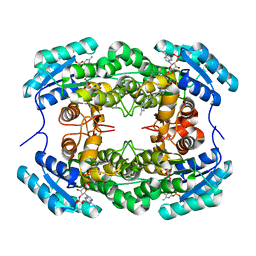 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | Descriptor: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Yin, D.J, Rao, Y.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
8HFK
 
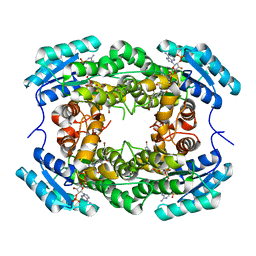 | |
7YB2
 
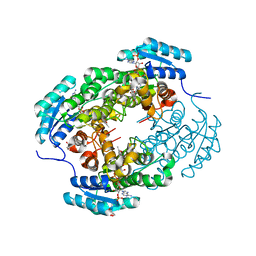 | | Crystal Structure of anthrol reductase (CbAR) in complex with NADP+ and emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
7YB1
 
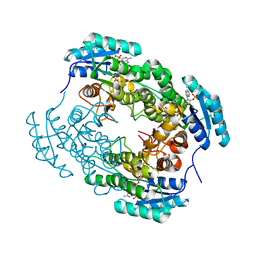 | | Crystal Structure of anthrol reductase (CbAR) in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
7Y3X
 
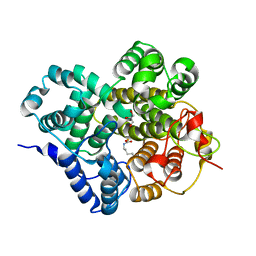 | | Crystal structure of BTG13 mutant (H58F) | | Descriptor: | FE (III) ION, GLYCEROL, Questin oxidase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3Y
 
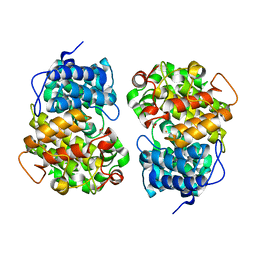 | | Crystal structure of BTG13 mutant (T299V) | | Descriptor: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | Authors: | Hou, X.D, Fu, K, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3W
 
 | |
7VM0
 
 | | Crystal structure of YojK from B.subtilis in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glycosyl transferase family 1, ... | | Authors: | Hou, X.D, Guo, B.D, Rao, Y.J. | | Deposit date: | 2021-10-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Front Bioeng Biotechnol, 10, 2022
|
|
5ZED
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5Z2X
 
 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5ZEC
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
8I6N
 
 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - L6T | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Ma, D, Cheng, Z.Y, Lai, Q.P, Hou, X.D, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2023-01-29 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Co-type nitrile hydratase mutant L6T from Pseudomonas thermophila at 2.2 Angstroms resolution.
To Be Published
|
|
7W8M
 
 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
