7MGO
 
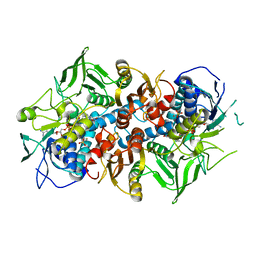 | | Crystal structure of F501H variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Prussia, G, Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
7MGN
 
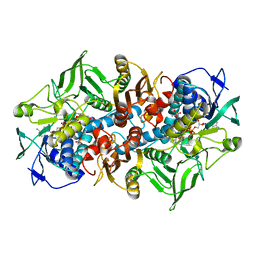 | | Crystal structure of F501H/H506E variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Zadvornyy, O.A, Prussia, G, Peters, J.W. | | Deposit date: | 2021-04-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
6CDK
 
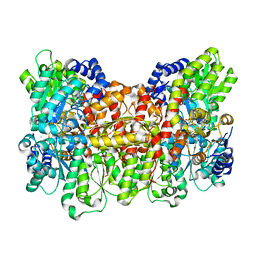 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
