1N0W
 
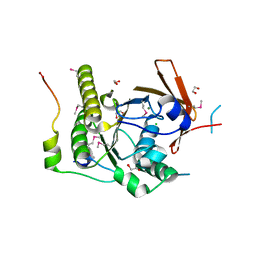 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
1SRS
 
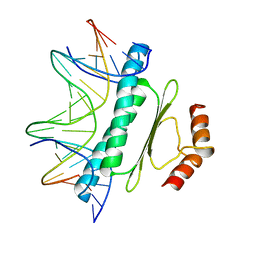 | | SERUM RESPONSE FACTOR (SRF) CORE COMPLEXED WITH SPECIFIC SRE DNA | | Descriptor: | DNA (5'-D(*CP*CP*(5IU)P*TP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*A P*AP*G)-3'), PROTEIN (SERUM RESPONSE FACTOR (SRF)) | | Authors: | Pellegrini, L, Tan, S, Richmond, T.J. | | Deposit date: | 1995-07-28 | | Release date: | 1995-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of serum response factor core bound to DNA.
Nature, 376, 1995
|
|
2ILR
 
 | |
1E0O
 
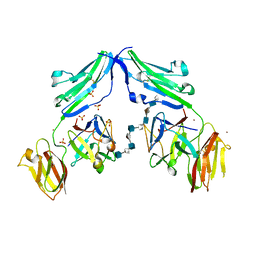 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
6T9Q
 
 | |
5OFN
 
 | |
5NWL
 
 | | Crystal structure of a human RAD51-ATP filament. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 1, MAGNESIUM ION | | Authors: | Pellegrini, L, Moschetti, T. | | Deposit date: | 2017-05-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA.
EMBO J., 37, 2018
|
|
5OF3
 
 | |
5OGS
 
 | | Crystal structure of human AND-1 SepB domain | | Descriptor: | MALONATE ION, WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Pellegrini, L, Simon, A.C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The human CTF4-orthologue AND-1 interacts with DNA polymerase alpha /primase via its unique C-terminal HMG box.
Open Biol, 7, 2017
|
|
4WWM
 
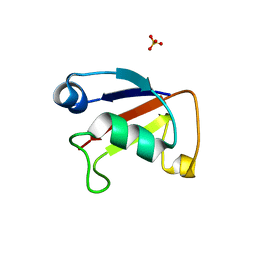 | | X-ray crystal structure of Sulfolobus solfataricus Urm1 | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Bray, S.M, Anjum, S.R, Blackwood, J.K, Kilkenny, M.L, Coelho, M.A, Foster, B.M, Pellegrini, L, Robinson, N.P. | | Deposit date: | 2014-11-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius.
Nat Commun, 6, 2015
|
|
5HOG
 
 | |
5HOI
 
 | |
6XTX
 
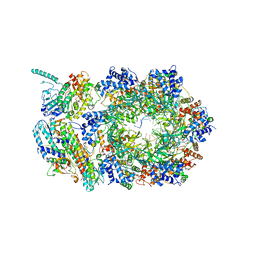 | | CryoEM structure of human CMG bound to ATPgammaS and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45 homolog, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rzechorzek, N.J, Pellegrini, L. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
3LGB
 
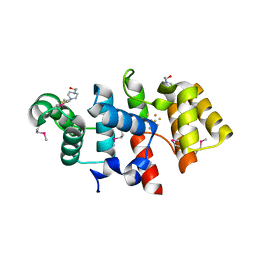 | |
7OPL
 
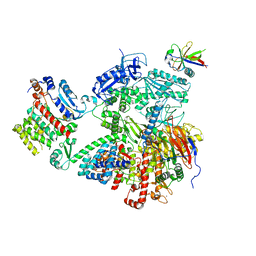 | |
6R4S
 
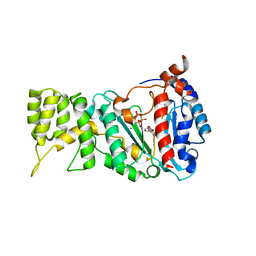 | |
6R4T
 
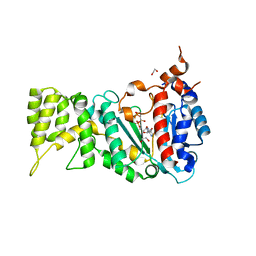 | |
6R5E
 
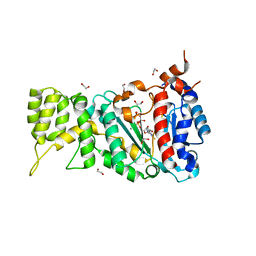 | |
6RB4
 
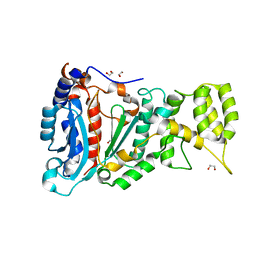 | |
6R5D
 
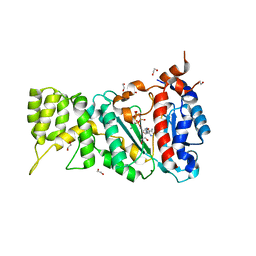 | |
6R4U
 
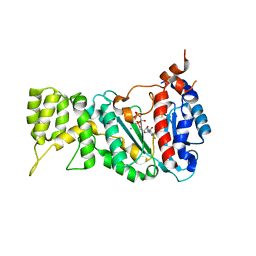 | |
7BGF
 
 | |
8BR2
 
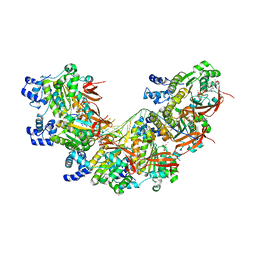 | | CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*CP*GP*AP*GP*CP*TP*CP*GP*AP*TP*GP*CP*AP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
8BSC
 
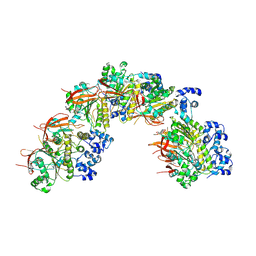 | |
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
